Part:BBa_K633001
Linker + EstA Membrane Protein
EstA is an outer membrane-anchored esterase from Pseudomonas aeruginosa. It has autotransporter activities that reside in its catalytic N-terminal domain, the C-terminal domain formes a β-barrel-like structure. EstA is inserted in the bacterial outer membrane where the N-terminal domain is translocated outside the outer membrane. (Becker, Theile, Heppeler & Michalczyk, 2005).
The successful cell-surface display of lipases as fusion proteins to an inactive variant of EstA has already been described. Passenger enzymes in that fusion retain their hydrolytic activities after being anchored on the outer surface of Escherichia coli cells. (Becker, Theile, Heppeler & Michalczyk, 2005)
An inactive EstA variant was used as an anchoring motif for the E. coli cell-surface display of lipolytic enzymes.
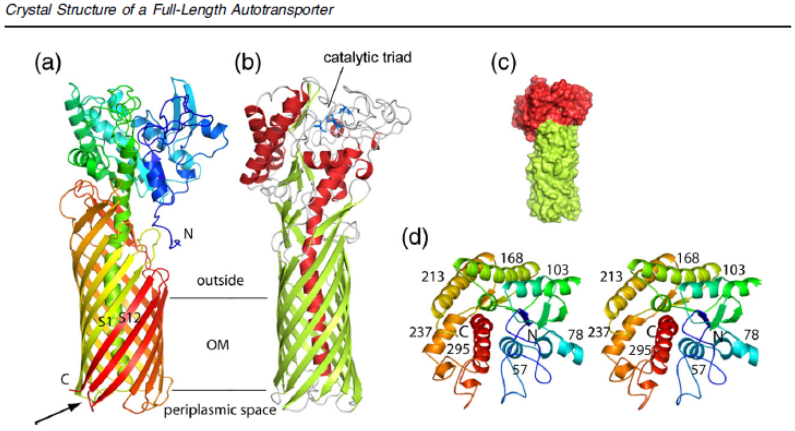
Fig. 1. Overall structure of the full-length EstA AT. (a) Backbone representation viewed from the side, with the protein
colored by a gradient from blue at the N-terminus to red at the C-terminus. Strands S1 and S12 are indicated, with an
arrow highlighting the connection between the central α-helix and strand S1. (b) Backbone view 90° rotated within the
plane of the membrane relative to (a), with helices colored red, β-strands colored green, and loops colored gray. The
catalytic triad residues are shown as blue stick models. The approximate location of the OM core is indicated by
horizontal lines. (c) Surface view of EstA from the side, with the β-barrel domain colored green and the passenger domain
colored red. (d) Stereo view of the EstA passenger from the extracellular side, colored as a rainbow from dark blue at the
N-terminus to dark red at the C-terminus. The numbers are those for the central residue of the α-helix. For clarity, the
loops have been smoothed. All figures were made using PyMOL.18
Three Dimensional Structure of the "EstA" Membrane Protein, including its esterease translocated domain.
We used only its transmembranal domain, without its functional esterease. This approach enables the exchange of different small proteins.
References:
Becker, S., Theile, S., Heppeler, N., & Michalczyk, A. (2005). A generic system for the escherichia coli cell-surface display of lipolytic enzymes. FEBS Letters, 579(5), 1177-1182. Retrieved from http://www.sciencedirect.com/science/article/pii/S0014579305000748
Van Den Berg, B. (2010). Crystal structure of a full-length autotransporter. Journal of Molecular Biology, 396(3), 627-633. Elsevier Ltd. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/20060837
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 217
- 1000COMPATIBLE WITH RFC[1000]
| None |
