Part:BBa_K4439001
N-[AS]4-C
Recombinant Green Lacewing Silk Protein
Contents
Abstract
For the scope of our project, we designed a new recombinant silk protein N[AS]4C which adds hydrophobic protective properties to the insulative cellulose aerogel we produced, when coated on top of it. This part is inspired from the N[AS]8C recombinant protein previously produced in E. coli BL21(DE3) (Part:BBa K4439002). This part has been designed but not built and tested, as for the project we used this basic part to design the composite part mSA-N[AS]4C-CBD-10xHis (Part:BBa K4439007), which we successfully built and tested.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Protein Characterization
- Silk proteins demonstrate interesting mechanical properties such as toughness, strength, lightweight, biodegradability and the possibility to produce different morphologies (fibers, foams, capsules, films). In addition to this, silk proteins comprise a high percentage of the amino acids glycine, serine and alanine which have an intermediate hydrophobicity.
- Green lacewing insects produce two types of silk: one produced by the larvae (cocoon) and the other by adult females (egg-stalk). The adult produced silk acts as a protective shelter and structural support for egg stalks, which are two ideal properties for a waterproof coating for our aerogel.
- In green lacewings, two serine- and glycine-rich silk proteins (Ma1XB1 and Ma1XB2) have been identified, both with highly repetitive core domains and small terminal domains. The core domain’s structure is rich in β-sheets with an approximative sheet-length of four amino acids between turns. These form repeating structural units constituting β-helices which have a significant positive correlation with the proteins’ surface hydrophobicity. A consensus motif for the core domain of Ma1XB2 (named [AS])had already been generated. Furthermore, a recombinant protein constituted by 8 repetitions of this [AS] module had also already been expressed in E. coli.
- From N[AS]8C recombinant protein, one can design another recombinant green lacewing silk protein with only 4 [AS] modules. It is smaller than N[AS]8C protein but should keep the same properties.
Modeling
Modeling the amount of proteins to coat a cellulose aerogel
We had different parameters that needed to be considered and we listed the most of them in (Table. 1)

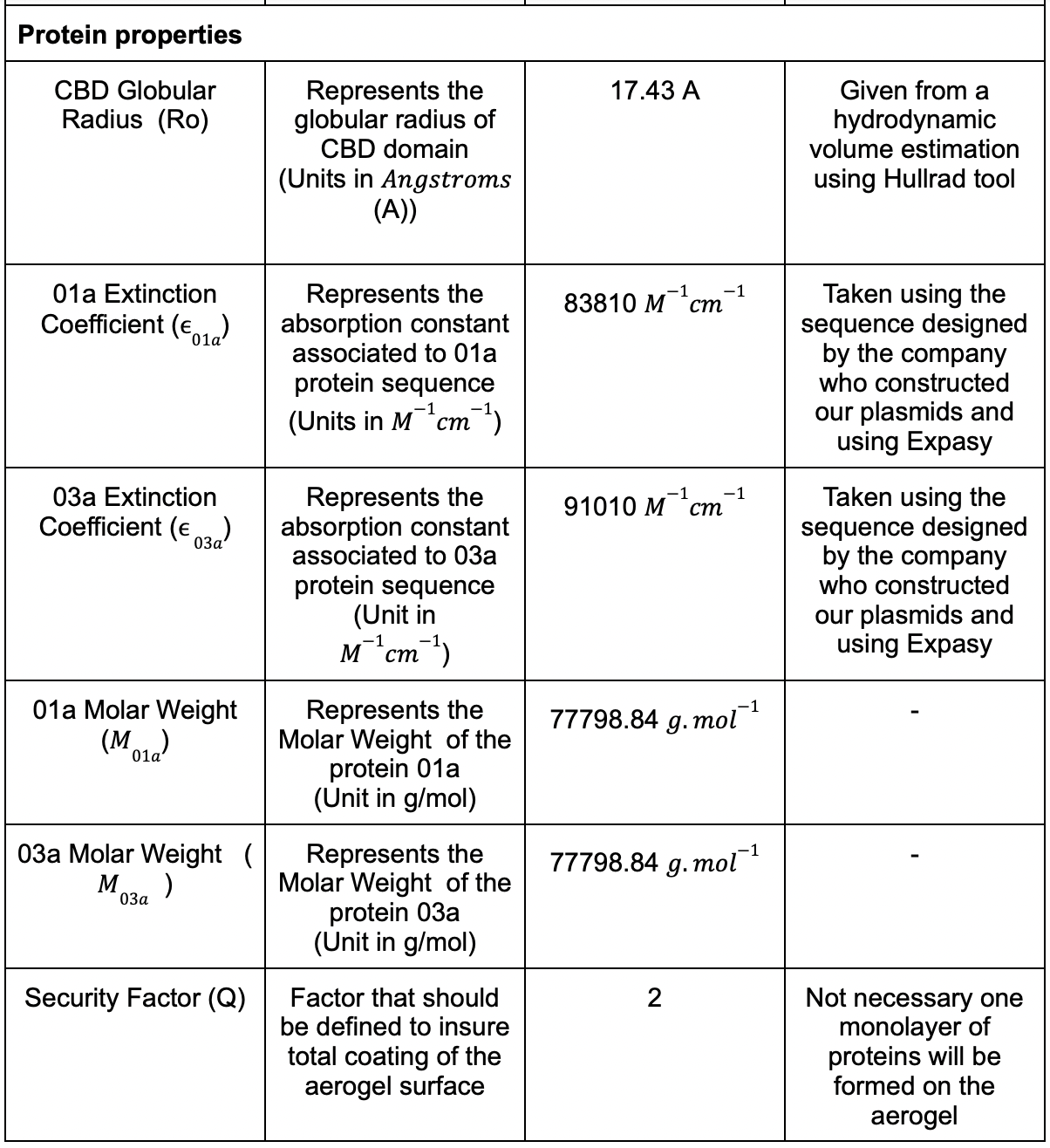
Table 1: Table for the parametric model .
In the proposed model we would like to coat geometrically our aerogel surface with different CBD globular entities. Since they are the most probable point of anchoring they will represent an entire protein in the counting of the amount of proteins. We supposed that the proteins would be placed in an adjacent manner.
First step was to consider the aerogel as a perfect cylinder that will be coated by the proteins. The surface of the cylinder is simplified to two circles and an elongated rectangle (fig2). We proceeded to a geometrical filling of the surface.

Figure 2 | Aerogel simplified representation as a perfect cylinder.
The attaching CBD region was taken from the above model 4JO5 to study its hydrodynamic characteristics using the Hullrad algorithm. This program was used to get specific useful information on the protein 4JO5:

Table 2 | Hullrad computation for CBD, AJO5
Using the values from (Table. 2), we populated the different surfaces of the aerogel with circular proteins using some simple geometry equations. The two values of AVSR and MD were studied in the example. Considering the protein radius and aerogel diameter, we came up with an iterative algorithm in three steps: Step 1: Populate the outer peripheral of the bigger cylinder:
We used the following angular equation to compute the angle of placement of the first small protein circle in the aerogel bigger circle:
alpha = 1 /π |arcsin(r(proteins)/(r(aerogel)- r(proteins)))*180| (1)
where r(proteins) and r(aerogel) represent respectively the radius of the proteins and the aerogel.
Step2: Compute the number of cylinders that can populate the peripheral of our the bigger circle:
nb(circles) =[360/(2*alpha)] (2)
where [x] is the greatest integer function s.
Step 3: Define a secondary circle that has the next small circles:
We iterated the new radius until we could not fit any small circles in the area (fig.12):
r(new)=r(previous)-( 2 *r(small)) (3)
where r(previous) is the large radius of the previous iteration.
Ending condition :
r(previous)< r(small)
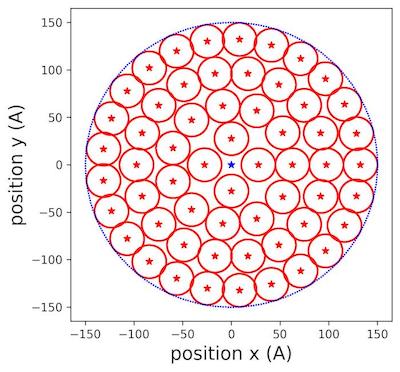
Figure 3 | Populating circles with tiny cylinders zoom at 150 Å aerogel surface .
In (fig.3), we saw clearly how the different circles were placed inside the allocated surface of aerogel. We extended the reasoning to filling the rectangular lateral surface.
For the lateral specific surface, the operation is described as following:
Step 1: Divide the width of the rectangle by the number of small protein circles that fits in the line :
nb(circles for width) = [W2 * r(proteins)] (4)
where W is the width of the rectangle, r(proteins) represents the radius of the proteins and [x] is the greatest integer function as described above.
Step 2: Multiply the above number by the number of lines that can fit in the height of the rectangle:
nb(circles for rectangle) = [h2* r(small)] * nb(circles for length) (5)
where h represents the height of the rectangle, [x] is the greatest integer function as described above.
By taking the parameters of the aerogel from (Table.1) and the parameters of the CBD from (Table.2) we got the following output in number of molecules and in number of molar:

Table 3: Results of the coating computations.
- The values found in (Table.3) were useful to define the protein solution volume that we soaked the hydrogels or the aerogels with.
References
- Bauer, F. & Scheibel, T. (2012, 16 may). Artificial Egg Stalks Made of a Recombinantly Produced Lacewing Silk Protein. Angewandte Chemie International Edition, 51(26), 6521‑6524. https://doi.org/10.1002/anie.201200591
- Bauer, F. (2013) Development of an artificial silk protein on the basis of a lacewing egg stalk protein. https://epub.uni-bayreuth.de/72/1/Diss.pdf
| None |
