Part:BBa_K4380018
eGFP and TA2 peptide fusion protein
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 151
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 820
Illegal SapI.rc site found at 16
Contents
Introduction
Vilnius-Lithuania Igem 2022 project NanoFind was working to create an easily accessible nanoplastic detection tool, using peptides, whose interaction with nanoplastic particles would lead to an easily interpretable response. The system itself focused on smaller protein molecules, peptides, which are modified to acquire the ability to connect to the surface of synthetic polymers – plastics. The detection system works when peptides and nanoplastic particles combine and form a sandwich complex - one nanoplastic particle is surrounded by two peptides, attached to their respective protein. The sandwich complex consisted of two main parts – one is a peptide bound to a fluorescent protein, and the other peptide is immobilized on a cellulose membrane by a cellulose binding domain.
This specific part is a green fluorescence reporter protein bound to a specific plastic binding TA2 mutated peptide fused by a linker.
Cultivation and purification
- medium: Luria Bertani (LB) medium
- strain: E.coli Arctic Express(DE3)
- antibiotics: 30 µg mL-1 Kanamycin
- temperature: 37 °C
- cultivation time: 16 h 16 °C
- pH of lysis buffer : 7.4
E.coli strain was transformed with plasmid expression vector pet29b(+) containing the desired GFP fusion protein. Bacteria night culture is grown, which after 16 h 1/30 dilution is sown into a larger volume of liquid LB medium with appropriate antibiotic (Kanamycin). Cell culture was grown at 37 ℃ at 200 rpm until OD600 value suitable for induction (0.5-0.6) is achieved. IPTG is then added to the growth medium to a concentration of 0,5 mM and the bacteria are further grown for 16 h at 16 °C. Cells are collected by centrifugation for 5 min. at 7000 rpm at 4 °C. (Figure 1)
Specific plastic binding
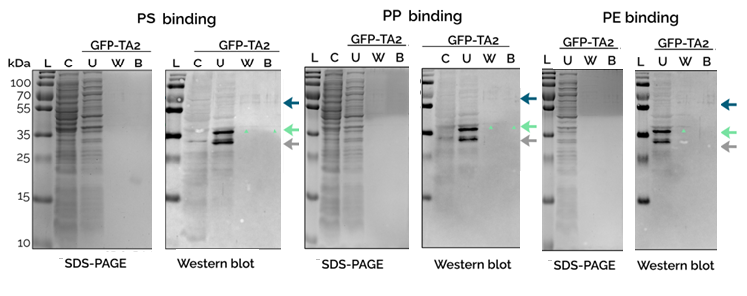
Proof that the peptide GFP-TA2 bind to plastics a specific plastic binding test was performed. Cell-free extracts were incubated with macro-sized pieces of plastics. After washing the unbound fraction, bound peptides were eluted. (Figure 2).
Western blotting (WB) was needed to see the faint bands due to the low concentration of peptides in the cell-free extract (Figure 2). The same cell-free extracts, whose profiles are shown in Figure 1, were used. The bands marked with green arrows were confirmed to be the GFP-TA2 peptides, as their intensity in WB compared to the whole protein profile in the same line of SDS-PAGE increased substantially. Bands seen in washed (W) and/or bound (B) fractions were considered to indicate protein binding. This way GFP-TA2 was confirmed to bind to PS, PP, and PE.
Binding to plastics - protocol
Peptide binding to plastics was tested using 3 ml glass bottles.
1. 20 mg of PS, PP, or PE was added to a glass bottle.
2. 300 µl of a cell-free extract was added into the bottle and the mixture was incubated for 30 min at room temperature, shaking at 300 rpm.
3. After incubation, a sample containing unbound proteins was taken for SDS-PAGE.
4. The leftover liquid was centrifuged through a hydrophilic PVDF membrane at 3200 rcf for 1 min.
5. 300 µl of 50 mM Tris-HCl, pH 7,4 was added and a sample containing washed proteins was taken for SDS-PAGE.
6. The leftover liquid was centrifuged through a hydrophilic PVDF membrane at 3200 rcf for 1 min.
7. 100 µl of 50 mM Tris-HCl, pH 7,4, 300 mM NaCl, 0,1 % Triton X-100 was added to elute peptides. The mixture was incubated for 30 min. at room temperature, shaking at 300 rpm.
8. After incubation, a sample containing bound peptides was taken for SDS-PAGE.
Due to low concentration of GFP-TA2 peptide, plastic binding was shown by Western blot additionally.
| None |