Part:BBa_K4345004
NarX mutant fused to mPapaya with a rigid linker
For more information on NarX, go to this part page: BBa_K4345000. The mutant was designed based on a study of Cavicchioli et al., (1995). The histidine on place 399 was replaced by glutamic acid. This allows the NarX mutants to dimerize but blocks it from phosphorylating NarL (the second messenger).
This composite part consists of BBa_K4345008 (NarX H399-E), BBa_K4345002 (Rigid linker) and BBa_K4345003 (mPapaya).
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 2153
Illegal PstI site found at 659 - 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 2153
Illegal PstI site found at 659 - 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 2153
Illegal XhoI site found at 260 - 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 2153
Illegal PstI site found at 659 - 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 2153
Illegal PstI site found at 659 - 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
This particular narX protein was derived from E. coli.
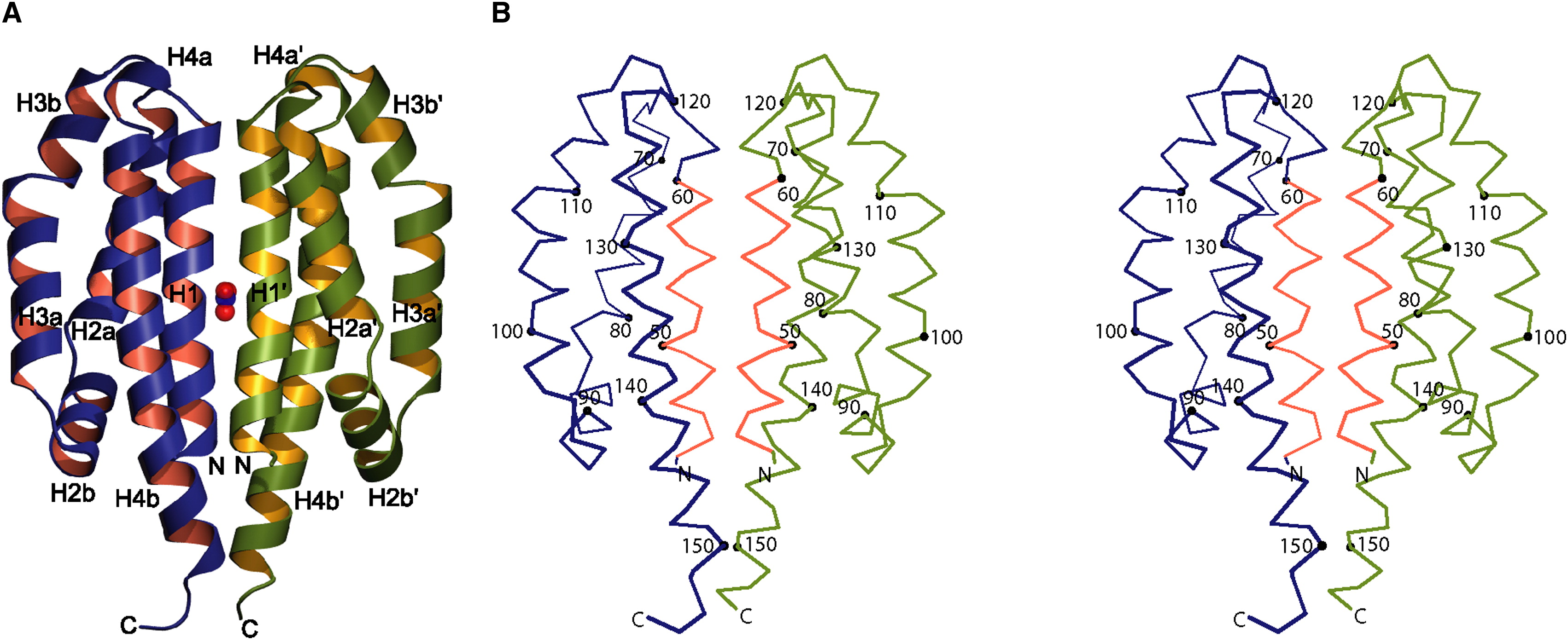
Image obtained from Cheung & Hendrickson, 2009
References
Cavicchioli, R., Schröder, I., Schröder, S., Constanti, M., & Gunsalus, R. P. (1995). The NarX and NarQ Sensor-Transmitter Proteins of Escherichia coli Each Require Two Conserved Histidines for Nitrate-Dependent Signal Transduction to NarL. JOURNAL OF BACTERIOLOGY, 177(9), 2416–2424.
Cheung, J., & Hendrickson, W. A. (2009). Structural Analysis of Ligand Stimulation of the Histidine Kinase NarX. Structure, 17(2), 190–201. https://doi.org/10.1016/J.STR.2008.12.013
narX sensor histidine kinase NarX [ Escherichia coli str. K-12 substr. MG1655 ]. (2022, September 22). National Library of Medicine - National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/gene/945788
//cds/membrane/receptor
| activation_coeficient | |
| protein | |
| signalling_molecule |