Part:BBa_K4211015
REDMAP-MazF
REDMAP system is a red light-activated system. The research team from East China Normal University reported a red/far-red light-mediated and miniaturized Δphytochrome A (ΔPhyA)-based photoswitch (REDMAP) system based on the plant photoreceptor PhyA in 2022. The system can rapidly bind the shuttle protein far-red elongated hypocotyl 1 (FHY1) under illumination with 660-nm light with dissociation occurring at 730 nm.
Based on the article, The BNUZH-China iGEM team 2022 designed a red light-activated system. We fused 5×UAS-P(hCMVmin) (BBa_K4211005) with the gene of interest. Therefore, we added a TAA (BBa_M36117) sequence to terminated the transcription of the gene of interest. At the C-terminal of the gene of interest, we added a CMV promoter (BBa_K1439007) to control the expression of the downstream target genes. In order to sense the red light, we bound PhyA (BBa_K4211007), Gal4 (BBa_K4211009), FHY1 (BBa_K4211011) and VP64 (BBa_K4211013) at the C-terminal of the CMV promoter. In land plants, the ability to sense and respond to red/far-red light depends on proteins of the phytochrome photoreceptor family. PhyA is a well-characterized member of this family that binds covalently to a chromophore known as Phycocyanobilin (PCB). PhyA is a red/far-red light-responsive photoreceptor from the plant Arabidopsis thaliana. The PCB binds covalent to PhyA to produce responses to red light (λmax = 660 nm) and far-red light (λmax = 730 nm). Following activation by red light, PhyA is transported into the nucleus based on interactions with shuttle proteins, including FHY1 and FHY1-like (FHL). The Gal4 transcription factor is a positive regulator of gene expression of galactose-induced genes. This protein represents a large fungal family of transcription factors, Gal4 family, which includes over 50 members in the yeast Saccharomyces cerevisiae. FHY1, the shuttle protein far-red elongated hypocotyl 1, can rapidly bind PhyA under illumination with 660 nm light and can dissociate occurring at 730 nm. When PhyA is interacting with FHY1, it can be transported into the nucleus. VP64, the herpes simplex virus-derived VP16 activation domain, is a kind of transcriptional activators. It is fused to FHY1. When illuminated with the 660 nm light, PhyA can bind with FHY1 and can be transported into the nucleus. According to the data given by the article, ΔPhyA–Gal4/FHY1–VP64 combination demonstrated the best switching performance than any other. So, we combined PhyA with Gal4 and linked FHY1 with VP64. Therefore, we used PCB to activate PhyA.
MazF is a stable non-specific ribonuclease encoded by Bacillus subtilis. It is a toxin protein in MazE-MazF, toxin-antitoxin module. It is used in conjungtion with MazE, which is also encoded by Bacillus subtilis, to provide a toxin-antitoxin kill switch in various stressful conditions. It is function as mRNA endonuclease.
In this module, we replaced GOI with MazF. We hoped that when illuminated with the 660 nm light, MazF can be transcription and translation. Then MazF works, killing the engineered cells. Figure 1 illustrates the detailed design of the whole device.
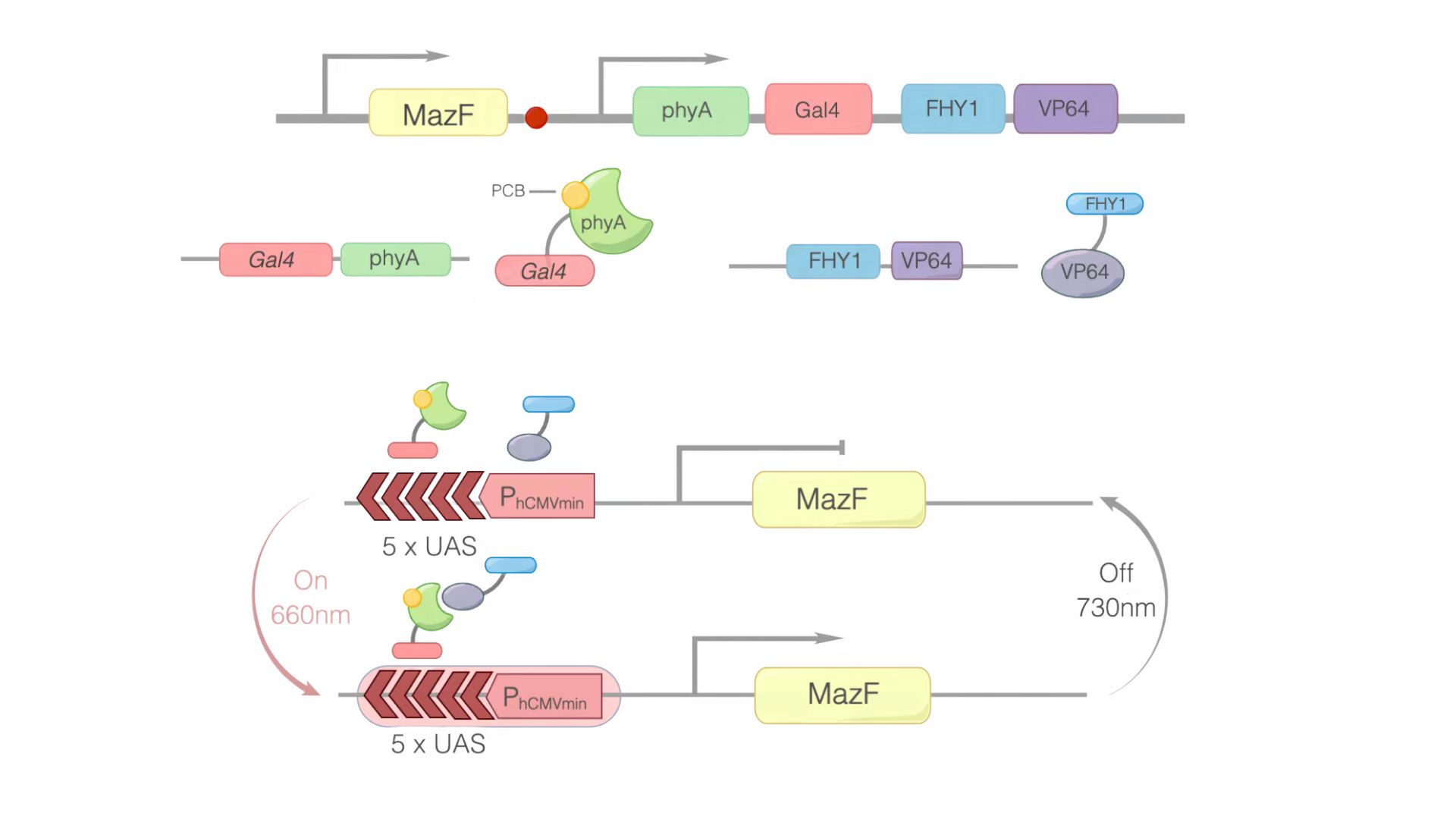
Figure 1: Construct design and the principle of the REDMAP-MazF. 5×UAS-P(hCMVmin) was fused to MazF. Gal4 was linked via a linker to PhyA. At the same time, PCB was combined with PhyA. PhyA turned to ΔPhyA. VP64 was linked with FHY1. When 660 nm light illuminating, ΔPhyA combines with VP64. Then, Gal4-ΔPhyA is transported into the nucleus by VP64-FHY1. After that, Gal4 will combine with 5×UAS-P(hCMVmin), activating the transcription of the downstream target gene: MazF. Then MazF works, killing the engineered cells.
Usage and Biology
We planned to use it to activate the transcription of MazF (BBa_K302033) to cause the engineered cells to commit suicide.
Experimental approach
For testing this device, we used HEK293T cells, which were seeded in 25cm2 flask. The PLVX vector was transiently transfected into HEK293T cells by Lipo8000™ transfection system and incubated for 43 h to ensure normal cell status. Then, phycocyanobilin (PCB) with a final concentration of 5 μM was added to the system, and the system was illuminated with red light (660 nm) for 3 h. After illumination, the cells were observed and the cell lysates were analyzed by SDS-PAGE and Western Blot.
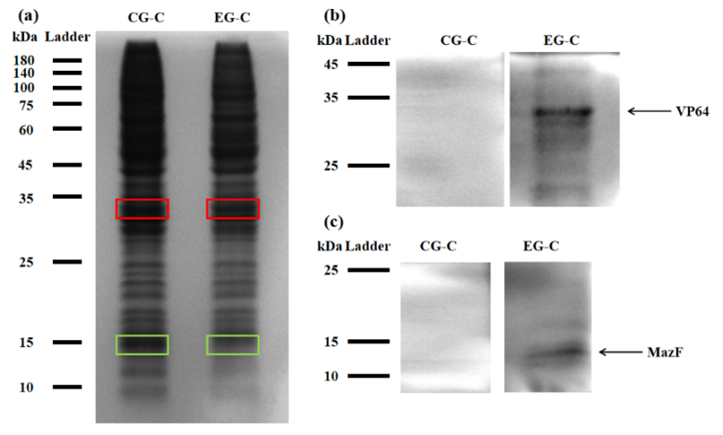
Figure 2: (a) REDMAP SDS-PAGE results. The VP64 strip is in the red box, and the MazF strip is in the green box. CG-C was the cell lysate of the control group, and EG-C was the cell lysate of the experimental group. (b) VP64 Western Blot diagram. CG-C was the cell lysate of the control group, EG-C was the cell lysate of the experimental group, and the theoretical size of VP64 was 30 kDa. (c) MazF Western Blot. CG-C was the cell lysate of the control group, EG-C was the cell lysate of the experimental group, and the theoretical MazF size was 13 kDa.
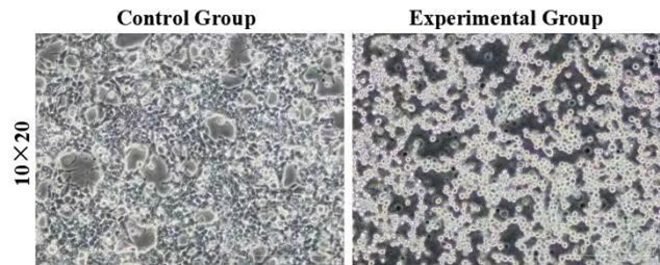
Figure 3: Morphology of control and experimental cells after red light verification. The control group was HEK293T cells without transient transfection, and the experimental group was HEK293T cells with transient transfection. 10×20 refer to the multiple of the eyepiece × the objective.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
//awards/part_collection
| None |
