Part:BBa_K4170020
LbuCas13a codon oprimized for E.coli
This plasmid contains the coding sequence (CDS) of codon-optimized LbuCas13a protein for recombinant LbuCas13a protein expression in E.coli. The genus Leptotrichia is the source of the LbuCas13a coding sequence. Cas13a is a type VI-A CRISPR-Cas RNA-guided RNA ribonuclease which degrades invasive RNAs targeted by CRISPR RNA (crRNA) and has potential applications in RNA technology.
Usage and biology
CRISPR/Cas13a systems as RNA sensors
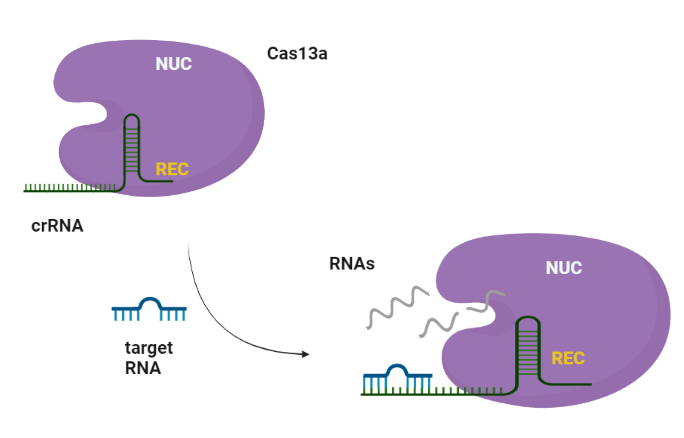
CRISPR-Cas systems are RNA-guided adaptive immune systems that protect prokaryotes from foreign genetic elements derived from evading viruses and phages (Shan et al., 2019). The Cas13a is ribonuclease with a double biological functionality: catalyzes the crRNA maturation and degrades the RNA-guided ssRNA (single-stranded RNA) interdependently using two separated catalytic sites (Rath et al., 2015). Generally, CRISPR/Cas system can be divided into two main classes, class I and II, according to the system comprising a single or multiple effectors (Liu et al., 2017). Among them, class II (e.g., Cas9, Cas12, and Cas13) possesses more widespread application, due to its simple components (a single effector protein and a programmable guide RNA, Wang et al., 2021). Cas13 can be further divided into four subtypes, Cas13a-d, exhibiting diverse primary sequences except the two highly conserved HEPN (higher eukaryotes and prokaryotes nucleotide-binding) domains, which are responsible for both cis- and trans-RNase activities (Florczuk et al., 2017). Structural studies revealed that Cas13a adopts a bilobed architecture including recognition (REC) and nuclease (NUC) lobes (Wang et al., 2021, Zhou et al., 2020). CRISPR-Cas12,13,14 exhibits nonspecific degradation of non target (trans cleavage) after specific recognition of nucleic acids, thus CRISPR/Cas biology promises rapid, accurate, and portable diagnostic tools, the next-generation diagnostics. Cas13a due to its propensity to cleave RNAs after binding a user-defined RNA target sequence, is used to detect single molecules of RNA species with high specificity (Liu et al., 2017).
Similarly to Cas9, Cas13a recognizes the short hairpin of the crRNA and made a complex. The target specificity is encoded by the 28-30-nt crRNA spacer sequence which is complementary to the target region. Cas13s exhibit collateral activity and after recognition and cleavage of the target transcript, degrades non-specifically any nearby RNAs regardless of complementarity to the spacer.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 3129
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 1207
- 1000COMPATIBLE WITH RFC[1000]
Citations
1. Kaminski, M., Abudayyeh, O., Gootenberg, J., Zhang, F. and Collins, J., (2021) "CRISPR-based diagnostics." Nature Biomedical Engineering, 5(7), pp.643-656.
2. Liu, L., Li, X., Ma, J., Li, Z., You, L., Wang, J., Wang, M., Zhang, X.and Wang, Y., (2017) "The Molecular Architecture for RNA - Guided RNA Cleavage by Cas13a." Cell, 170(4), pp .714 - 726.e10.2.
3. Rath, D., Amlinger, L., Rath, A. and Lundgren, M., (2015) "The CRISPR-Cas immune system: Biology, mechanisms and applications." Biochimie, 117, pp.119-128.
4. Shan, Y., Zhou, X., Huang, R.and Xing, D., (2019) "High - Fidelity and Rapid Quantification of miRNA Combining crRNA Programmability and CRISPR / Cas13a trans - Cleavage Activity." Analytical Chemistry, 91(8), pp .5278 - 5285. 3.
5. Wang, B., Zhang, T., Yin, J., Yu, Y., Xu, W., Ding, J., Patel, D. and Yang, H., (2021) "Structural basis for self-cleavage prevention by tag:anti-tag pairing complementarity in type VI Cas13 CRISPR systems." Molecular Cell, 81(5), pp.1100-1115.e5
6. Zhang, C., Konermann, S., Brideau, N., Lotfy, P., Wu, X., Novick, S., Strutzenberg, T., Griffin, P., Hsu, P.and Lyumkis, D., (2018) "Structural Basis for the RNA - Guided Ribonuclease Activity of CRISPR - Cas13d." Cell, 175(1), pp .212 - 223.e17.
| None |