Part:BBa_K4170017
LbuCas13a coding device under T7 promoter
This plasmid contains all the basic genetic parts required for the bacterial expression of codon-optimized LbuCas13a protein without the small ubiquitin-like modifier (SUMO) protein. Due to the absence of SUMO protein is observed enhanced expression of the LbuCas13a protein in the inclusion bodies (IBs) of the bacterial compared to the LbuCas13a-SUMO protein since SUMO favors the proteins’ solubility.
Usage and Biology
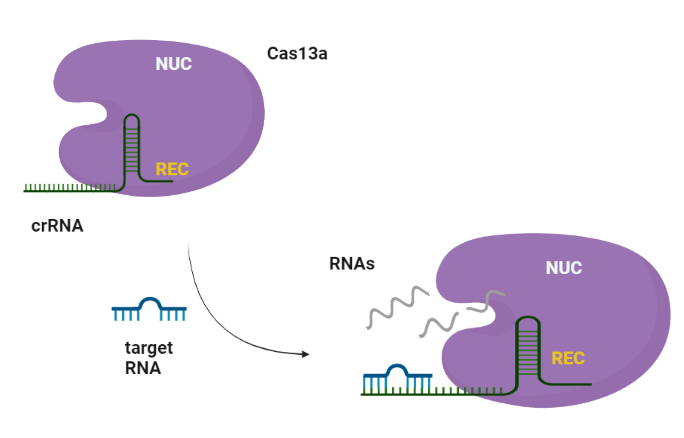
CRISPR-Cas systems are RNA-guided adaptive immune systems that protect prokaryotes from foreign genetic elements derived from evading viruses and phages (Shan et al., 2019). The Cas13a is ribonuclease with a double biological functionality: catalyzes the crRNA maturation and degrades the RNA-guided ssRNA (single-stranded RNA) interdependently using two separated catalytic sites (Liu et al., 2017).
Similarly to Cas9, Cas13a recognizes the short hairpin of the crRNA and made a complex. The target specificity is encoded by the 28-30-nt crRNA spacer sequence which is complementary to the target region. Cas13s exhibit collateral activity and after recognition and cleavage of the target transcript, degrades non-specifically any nearby RNAs regardless of complementarity to the spacer.
This innovative molecular system has provided breakthrough opportunities for RNA detection for clinical diagnostics. Since RNA molecules are key mediators in all biological processes, served as significant biomarkers to the clinical verification and differential diagnosis of cancer. MiRNAs are a class of small non-coding RNAs that participate in numerous cellular processes such as cellular development, proliferation and apoptosis. Indeed, deviating miRNA expression is often related to the occurrence of diseases such as cancer. Therefore, the accurate detection of differences in miRNAs detection serve could serve as significant biomarkers for cancer verification and diagnosis. The RNA detection approaches can be characterized into 2 main categories, the direct detection and the indirect nucleic acid amplification (NAA)-based detection. Both approaches have advantages but also significant disadvantages which hinder their wide application in clinical practice. The direct classical methods including Nothern blotting, microarrays and FISH, despite their high fidelity, lack high sensitivity and are labor-intensive, time-consuming and require specialized personnel. On the other side, NAA-based detection approaches such as reverse transcription PCR (RT-PCR) and other isothermal amplification techniques show high sensitivity but introduce additional steps such as the conversion of RNA to DNA and auxiliary DNA template replication steps in order to achieve signal amplification. These additional steps often result in considerable sample loss due to incomplete RT step, amplification bias caused by error-prone sequence amplification, and a high risk of false-positive results due to possible contamination. Furthermore, the requirement of thermocycler for the reactions, limit its application as a point of care medical diagnostic device. These issues render the existing IVD tools for cancer diagnostics inadequate for clinical application in routine base (Shan et al., 2019).
This genetic device is a part of the meta-CRISPR part collection developed by iGEM22_Thessaloniki_Meta.
Meta-CRISPR part collection
The meta-CRISPR part collection was developed in accordance with standardization, modularity and standard assembly rules aiming to enhance the CRISPR/crRNA applicability in synthetic biology projects. This collection allows the interchangeability of the genetic parts and the automation of construction following a standardized Golden Gate-based cloning strategy. Utilizing the meta-CRISPR collection, future iGEM teams can select the desired combination of DNA parts depending on the specific application of the CRISPR method (https://2022.igem.wiki/thessaloniki-meta/part-collection). Through the combination of different promoters and LbuCas13a coding sequences, one can select the desired LbuCas13a expression system, for example under the transcriptional control of the T7 or the pRha promoter. In addition, by combining the suitable genetic parts, researchers can select the purification method of the recombinant LbuCas13a protein either from inclusion bodies or from the soluble cytoplasmic fraction of the bacteria. Regarding the crRNA, utilizing the guidelines provided on the crPrep-crRNA preparation kit one can easily in silico design and produce the necessary crRNA sequence depending on the target miRNA utilizing the standardized cloning method and requiring only one additional primer. Last but not least, the LbuCas13a coding device can be assembled with the crRNA transcription system in a single plasmid enabling the simultaneous production of the LbuCas13a/crRNA complex in bacteria.

Basic parts assembled for device construction
The part sample which is flanked at the beginning and the end with prefix and suffix respectively, is composed of the following basic parts assembled together in series and downstream of the prefix:
- Bacterial terminator for LacI CDS (upstream of prefix): putative bacterial transcription terminator
- Biobrick Prefix sequence: BioBrick prefix for parts that do not start with "ATG"
- LacI Coding sequence: Lac repressor
- LacI promoter: Promoter that has the transcriptional control of Lac repressor
- T7 promoter: promoter for bacteriophage T7 RNA polymerase
- Lac operator: Lac repressor protein binding site
- Ribosome binding site: efficient ribosome binding site from bacteriophage T7 gene 10 (Olins and Rangwala, 1989)
- 6XHis tags: 6xHis affinity tag
- Thrombin site: thrombin recognition and cleavage site
- LbuCas13a CDS: Cas13a protein derived from Leptotrichia Buccalis
- Biobrick Suffix sequence : universal suffix for all parts
- His operon terminator (downstream of Suffix): putative transcription terminator from the E. coli
Cloning strategy and results
For the final assembly of the PCR amplified genetic elements into pSB1C3 plasmid we followed the Golden Gate-based ‘SevaBrick Assembly’ method and the SEVA 3.1 platform. As for all our final composite parts, the initial steps of our cloning strategy constitute the PCR amplification of different genetic elements, followed by the efficient assembly of the PCR amplified genetic products into pSB1C3 backbone. Further information are provided on the design page.
Step 1
- PCR amplification with SUMOLESS FWD and Cas13a P4 RVS primers using the final T7-SUMO-LbuCas13a as a template. This PCR produces the SUMOLESS-Cas13a part ready for Golden Gate assembly.
- PCR amplification with T7 P0 FWD and SUMOLESS RVS primers using the final T7-SUMO-LbuCas13a as a template. This PCR produces the SUMOLESS-T7 ready for Golden Gate assembly.
- PCR amplification with Ev and Pv standard primers from Basic SevaBrick Assembly [seva 3.1] using the Bba_J364007 part of the 2022 DNA distribution Kit. This PCR produced the pSB1C3 backbone linearized and ready for Golden Gate assembly.

PCR amplification was performed utilizing two pairs of primers and then the samples were loaded and run in 1 % agarose gel electrophoresis. As shown in figure 1 both PCR amplifications succeeded. On the left side the SUMOLESS-Cas13a part (3521 bp) is depicted and on the right side SUMOLESS-T7 part (1640 bp). The linearized pSB1C3 backbone amplified by PCR is depicted on figure 2.
Step 2
Golden Gate assembly of the PCR amplified SUMOLESS-Cas13a and SUMOLESS-T7 parts with the linearized pSB1C3 vector for the efficient construction of the SUMOLESS-LbuCas13a coding sequence under the transcriptional control of the T7 promoter.
The products of the Golden Gate assembly underwent transformation into E.coli DH5a competent cells and then colony PCR was performed, using VR and VF2 primers. For the colony PCR procedure, from the agar plate half amount of each colony was picked and diluted on 20 μl of dH20. The other half amount was picked for liquid culture. The samples were loaded and run in 1% agarose gel electrophoresis and then we concluded that the Golden Gate assembly was successful. As depicted in figure 3 from all colonies the SUMOLESS Cas13a (5395 bp) has been amplified.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1453
Illegal BglII site found at 4750 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1421
Illegal AgeI site found at 2828 - 1000COMPATIBLE WITH RFC[1000]
Citations
1. Liu, L., Li, X., Ma, J., Li, Z., You, L., Wang, J., Wang, M., Zhang, X.and Wang, Y., (2017) "The Molecular Architecture for RNA - Guided RNA Cleavage by Cas13a." Cell, 170(4), pp .714 - 726.e10.2.
2. Shan, Y., Zhou, X., Huang, R.and Xing, D., (2019) "High - Fidelity and Rapid Quantification of miRNA Combining crRNA Programmability and CRISPR / Cas13a trans - Cleavage Activity." Analytical Chemistry, 91(8), pp .5278 - 5285. 3.
| None |