Part:BBa_K4170008
Lbucas13a assembly part 1
This composite part contains the first of the 4 total parts required for the efficient assembly of the LbuCas13a complete CDS with the linealized pSB1C3 backbone following th Golden Gate-based SevaBrick assembly methodology.
Cloning strategy
For the final assembly of the PCR amplified genetic elements into pSB1C3 plasmid we followed the Golden Gate-based ‘SevaBrick Assembly’ method and the SEVA 3.1 platform (Damalas et al., 2020).
'SevaBrick assembly' method
The Golden Gate-based ‘SevaBrick Assembly’ method was introduced by Stamatios G. Damalas and colleagues (Damalas et al., 2020). This method consists of standardized primers and protocols and facilitates the straightforward one-step assembly of multiple genetic elements into the SEVA 3.1 or the pSB#X# (# is determined by the identity of the replication origin and the letter X is determined by the antibiotic resistance marker) backbones in a fast and reliable process. The SevaBrick Assembly is a method where all the parts and backbones to be assembled are PCR amplified from any SEVA 3.1 or BioBrick vector, using a core set of standard long primers. All SevaBrick primers anneal on standard sequences of the SEVA 3.1 or BioBrick vectors, introducing BsaI recognition sites for directional multipart assembly via Golden Gate (Figure 2).
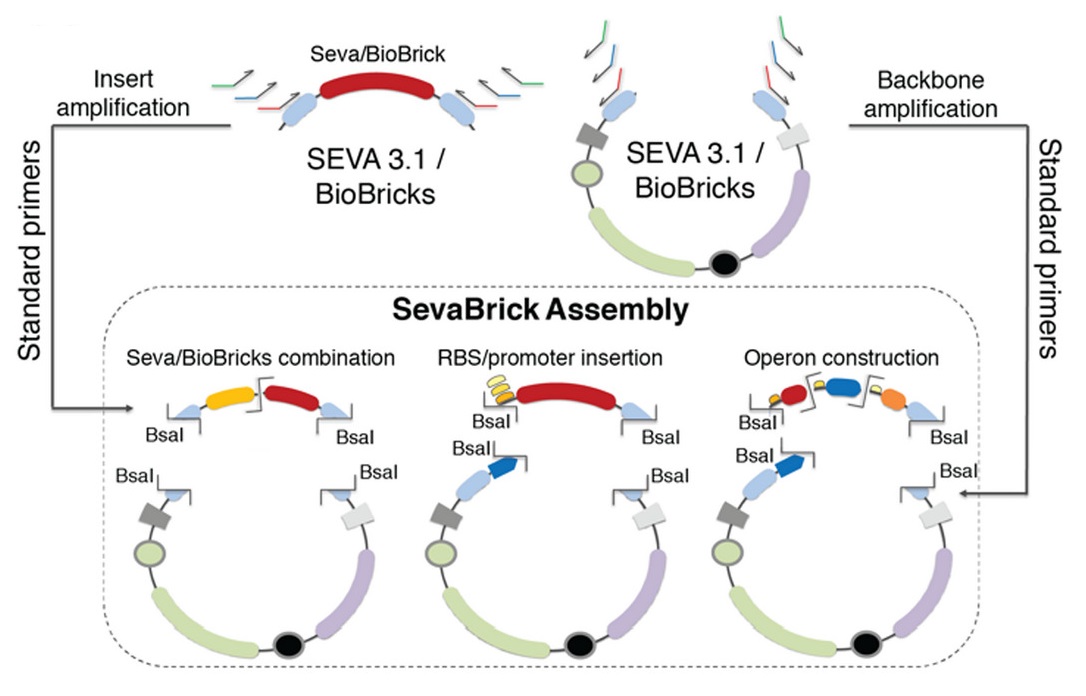
As for all our final composite parts, the initial steps of our cloning strategy constitute the mutagenesis PCR amplification of different genetic elements, followed by the efficient assembly of the PCR amplified genetic products into pSB1C3 backbone. The cloning process is described in detail below:
Step 1
- Mutagenesis PCR amplification with Cas13a P1 FWD and Cas13a P1 RVS primers using the pGJK_His-SUMO-LbuCas13a as a template. This PCR produces the P1 part ready for Golden Gate assembly.
- Mutagenesis PCR amplification with Cas13a P2 FWD and Cas13a P2 RVS primers using the pGJK_His-SUMO-LbuCas13a as a template. This PCR produces the P2 part ready for Golden Gate assembly.
- Mutagenesis PCR amplification with Cas13a P3 FWD and Cas13a P3 RVS primers using the pGJK_His-SUMO-LbuCas13a as a template. This PCR produces the P3 part ready for Golden Gate assembly.
- Mutagenesis PCR amplification with Cas13a P4 FWD and Cas13a P4 RVS primers using the pGJK_His-SUMO-LbuCas13a as a template. This PCR produces the P4 part ready for Golden Gate assembly.
- PCR amplification with Ev and Pv standard primers from Basic SevaBrick Assembly [seva 3.1] using the Bba_J364007 part of the 2022 DNA distribution Kit. This PCR produced the pSB1C3 backbone linearized and ready for Golden Gate assembly.
After the completion of the mutagenesis PCR amplifications, 1% agarose gel electrophoresis was conducted. As shown in Figure 1 all the PCR amplifications succeeded. Cas13a P1 part (1240 bp), Cas13a P2 part (734 bp) , Cas13a P3 part (1338 bp) , Cas13a P4 part (646 bp) and pSB1C3 backbone linearized (2052 bp) are depicted in Figure 1.

Step 2
Golden Gate assembly of the PCR amplified P1, P2, P3, P4 parts with the linearized pSB1C3 vector for the efficient construction of the SUMO-LbuCas13a coding sequence into pSB1C3 plasmid.
The products of the Golden Gate assembly underwent transformation into E.coli DH5a competent cells. To verify the successful Golden Gate assembly, colony PCR was performed using the primers VR and VF2. Then the samples were loaded and run in 1 % agarose gel electrophoresis. As depicted on Figure 2 from all colonies the desired SUMO-LbuCas13a (5908 bp) part has been amplified. For the colony PCR procedure, from the agar plate half amount of each colony was picked and diluted on 10 μl of dH20 performing the template DNA of the colony PCR. The other half amount was picked for the overnight liquid culture.

Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal prefix found in sequence at 9
- 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 9
Illegal NotI site found at 15 - 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 9
- 23INCOMPATIBLE WITH RFC[23]Illegal prefix found in sequence at 9
- 25INCOMPATIBLE WITH RFC[25]Illegal prefix found in sequence at 9
Illegal XbaI site found at 24 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 2
Illegal BsaI.rc site found at 937
Citations
1. Damalas, S., Batianis, C., Martin-Pascual, M., Lorenzo, V. and Martins dos Santos, V., (2020) "SEVA 3.1: enabling interoperability of DNA assembly among the SEVA, BioBricks and Type IIS restriction enzyme standards." Microbial Biotechnology, 13(6), pp.1793-1806.
| None |
