Part:BBa_K4162005
Hammerhead ribozyme

Fudan iGEM 2023
This is an effective self-cleaving ribozyme. In 2022, we used this ribozyme for the splicing of polycistron mRNAs and constructed ribozyme-assisted polycistronic co-expression system:pRAP based on this functionality.
In this year's experiments, we observed degradation of monocistronic mRNA after ribozyme cleavage, as reported in the original pRAP paper[1], affecting our functional characterization. Therefore, we added the stem-loop (liu2023) at the 3' end of the CDS to prevent mRNA degradation. We also design synthetic stem-loops using our software, with different strength, and experimental tested their anti-degradation capabilities in stem-loop test BBa_K4765129.
Simultaneously, we also constructed various ribozymes into our ribozyme test: leaky expression BBa_K4765120.
Introduction
Hammerhead ribozyme was first found in the genome of viruses and viroids. It involved in the processing of RNA transcripts based on rolling-circle replication. The tandem copy of RNA sequence will be generated in the roll ring replication, and the self-cleaving activity of ribozyme can ensure the generation of RNA copy of unit length.[2]
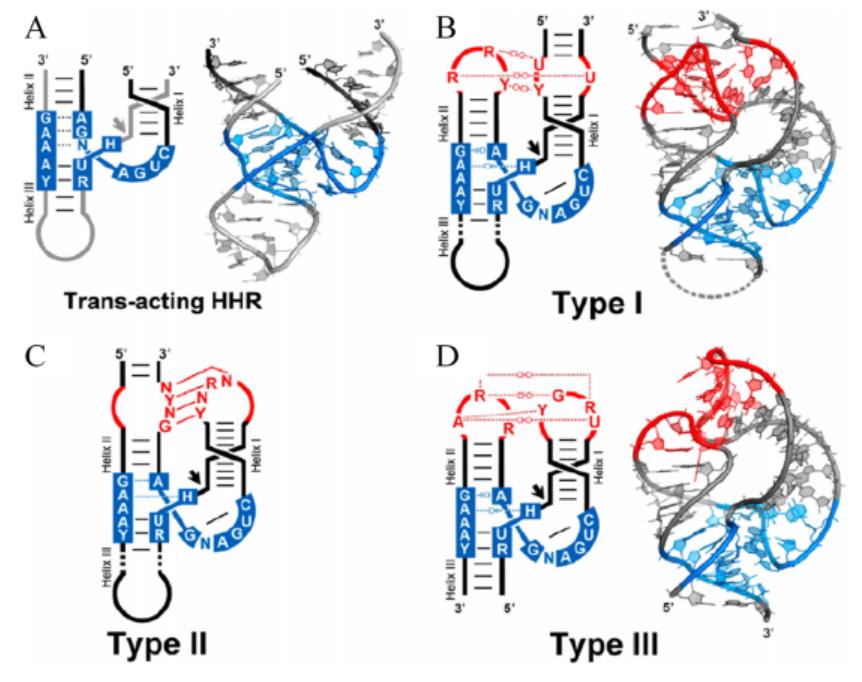
The secondary structure of hammerhead ribozyme resembles a hammer. According to different open helix tips, hammerhead ribozymes can be divided into three types: TypeⅠ, Type II and Type III. The catalytic center of ribozyme consists of 15 highly conserved bases surrounded by three helixs(HelixⅠ, Helix II and Helix III). The long-range interaction between HelixⅠ and Helix II can help stabilize the conformation of the catalytic center of the enzyme and improve the catalytic efficiency (Figure 1).[3] While working, ribozymes utilize a network of defined hydrogen bonds, ionic and hydrophobic interactions to generate catalytic pockets, which capitalize on steric constraints to generate in-line cleavage alignments and general acid-base chemistry to catalyze site-specific cleavage of the phosphodiester backbone.[4]
Contents
Usage and Biology
We construct a ribozyme-assisted polycistronic co-expression system (pRAP) by inserting ribozyme sequences between CDSs in a polycistron. In the pRAP system, the RNA sequences of hammerhead ribozyme conduct self-cleaving, and the polycistronic mRNA transcript is thus co-transcriptionally converted into individual mono-cistrons in vivo. Self-interaction of the polycistron can be nullified and each cistron can initiate translation with comparable efficiency. Besides, we can precisely manage this co-expression system by adjusting the RBS strength of individual mono-cistrons. In our project, we used ribozyme to build our crtYEBI and crtEBIY biobricks.
Characterization
Successful protein expression
As many researches indicate, the major problem of polycistronic vectors, which contain two or more target genes under one promoter, is the much lower expression of the downstream genes compared with that of the first gene next to the promoter[5]. The tail of the preceding coding sequence (CDS) can interfere with the head of the ribosome binding site (RBS), which can hinder RBS from combining to ribosomes. Such shortage occurred when we assembled crtEBIY without flanking ribozyme sequentially only to find incomplete expressions of our target proteins.
We first assemble four basic modules: BBa_K4162010(ribozyme + T7_RBS + crtE), BBa_K4162013(ribozyme + T7_RBS + crtB), BBa_K4162016(ribozyme + T7_RBS + crtI) and BBa_K4162019(ribozyme + T7_RBS + crtY). Next, we apply overlapping PCR to assemble composite modules which contain 2-3 ribozyme+RBS+CDS modules. Finally, we obtained several biobricks composed of four basic modules assembled in different orders. SDS-PAGE showed that the proteins in each module were successfully expressed and were intact proteins rather than short peptides after incomplete expression.


Stable expression among clones
For the composite modules constructed using the ribozyme, the self-interaction of polycistron is avoid. Our ribozyme promotes cleavage of transcribed mRNA and separated mRNA contains ribozyme+RBS+CDS just enough for ribosomes to translate.
When using ribozyme-assisted pRAP, the difference between clones is minimal. As shown in Figure 4 and 5, different clones of E. coli transformed with the same plasmid/construct were cultured using 96-well plates. After shaking culture overnight, the plate was spin at 4500 rpm for 30 minutes and bacteria was pelleted to the bottom of the wells. There is no clonal variation, showing stable expression of the individual mono-cistrons separated by ribozyme sequences (i.e., no trucated expression nor null function protein of downstream genes due to being placed under one promoter).

Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
References
- ↑ Liu Y, Wu Z, Wu D, Gao N, Lin J (2023). Reconstitution of Multi-Protein Complexes through Ribozyme-Assisted Polycistronic Co-Expression. ACS Synth Biol, 12(1), 136-143. https://doi.org/10.1021/acssynbio.2c00416
- ↑ Ferré-D'Amaré, A. R., & Scott, W. G. (2010). Small self-cleaving ribozymes. Cold Spring Harbor perspectives in biology, 2(10), a003574. https://doi.org/10.1101/cshperspect.a003574
- ↑ Jimenez, R. M., Polanco, J. A., & Lupták, A. (2015). Chemistry and Biology of Self-Cleaving Ribozymes. Trends in biochemical sciences, 40(11), 648–661. https://doi.org/10.1016/j.tibs.2015.09.001
- ↑ Ren, A., Micura, R., & Patel, D. J. (2017). Structure-based mechanistic insights into catalysis by small self-cleaving ribozymes. Current opinion in chemical biology, 41, 71–83.
- ↑ Kim, K. J., Kim, H. E., Lee, K. H., Han, W., Yi, M. J., Jeong, J., & Oh, B. H. (2004). Two-promoter vector is highly efficient for overproduction of protein complexes. Protein science: a publication of the Protein Society, 13(6), 1698–1703.
| None |