Part:BBa_K4146502
aroGfbr
aroG, as a metabolic pathway gene that has attracted much attention for many years, has always been a worthy object of study. We believe that its feedback inhibition may affect the synthesis of 2-phenylethyl alcohol. We use this Mutant Feedback-Resistant Version of aroG for resisting feedback inhibition and improving enzyme activity to enhance the synthesis pathway in the de novo synthesis of 2-phenylethyl alcohol.
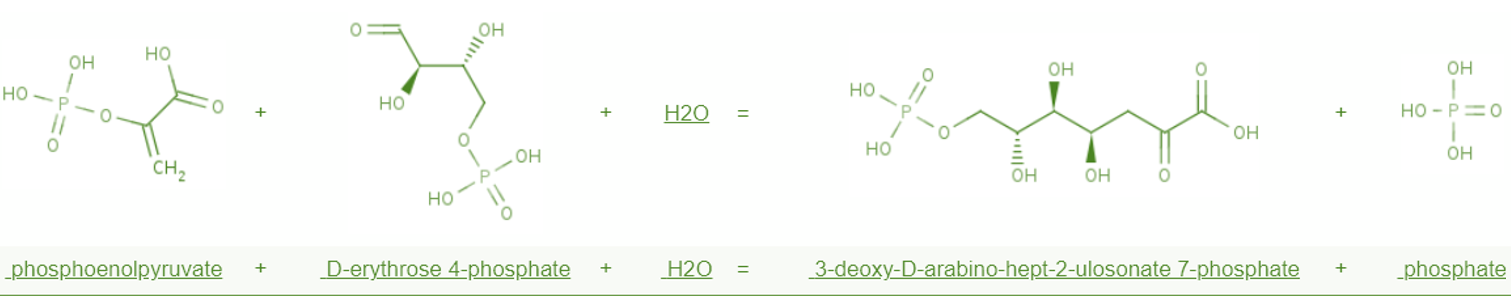
Figure 1. Biochemical reactions catalyzed by aroG
This gene encodes an enzyme that catalyzes the synthesis of DAHP, which directs carbon flow to the phenylalanine synthesis pathway. The phenylalanine-bound hydrophobic pocket consists of 12 amino acid residues, including Asp6, Asp7, Ile10, Pro150, Gln151, Leu179, Ser180, Phe209, Ser211 and Val221.
By reading the literature, we carefully interpreted the reasons behind the inhibitory effect of phenylalanine on aroG catalysis.
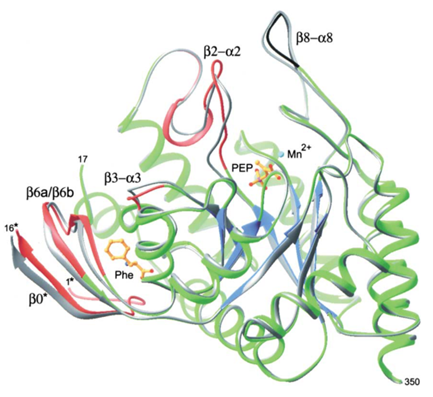
Figure 2. Structural changes in the DAHPS functional domain upon Phe binding. The protein chain (green, blue, red, black), Mn2+(cyan), phosphoenolpyruvate , and Phe (both in gold) are shown for +Phe DAHPS. The superimposed main chain of the -Phe enzyme is shown in gray . The main chains of four segments colored in red shift significantly upon Phe binding. These four segments are involved in transmission of the inhibition signal within a subunit from the Phe binding site to the active site. The change in conformation of the β8-α8 loop segment, shown in black, is due to differences in crystallization conditions.
When phenylalanine inhibits aroG, the crystal structure of the enzyme will undergo a large conformational change. The binding of Phe triggers the breaking and formation of many contacts between residuesIn this enzyme, the solvent enters the cavity of the binding site. Upon Phe binding, the entire N-terminal fragment of the constituent subunit of the compact dimer becomes ordered, forming a lid that participates in the coordination of the bound Phe and insulates its portion from solution.
The binding of Phe initiates the breaking and formation of many contacts between residues. They bind to each other to form many new hydrogen bonds, and these new interactions bind different segments of the polypeptide chain of the aroG protease together, allowing the enzyme to emerge in a new conformation.
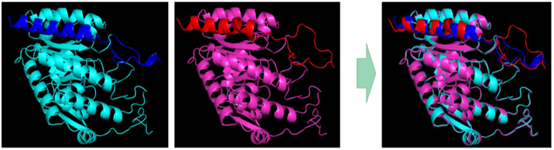
BUCT modeled aroG-fbr homology and aligned the molecular model of the enzyme with the original type BBa_K1060000:
Figure 3.BUCT homology modeling aroG-fbr, original type on the left is blue and BUCT mutant on the right is pink.
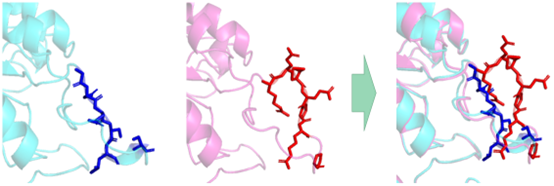
We hypothesized that the conformational change of the enzyme protein caused by the mutation might be the main reason for weakening the inhibitory effect of Phe. In the model comparison results, we found that the mutated molecular pocket appears to be sealed by a larger lid.
Figure 4. The red and dark blue sections represent the molecular conformational changes after the aroG sequence mutation. The spatial position of residues in front of the molecular cavity increases.
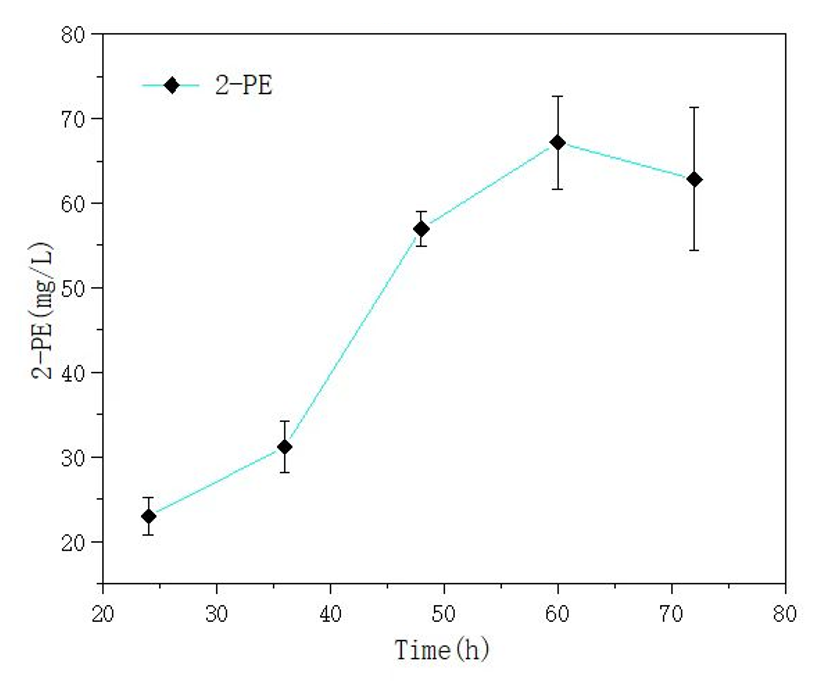
We also used this mutant gene to synthesize 2-phenylethyl alcohol (2-PE), and the yield results can be seen in the following figure. In our fermentation experiment, the highest yield of 2-PE reached 67.16mg/L.
Figure 5. yield of 2-PE in E. coli Nissle 1917
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |