Part:BBa_K4096006
xynA-PKC-OP
xynA-PKC-OP
Profile
Name: xynA-PKC-OP
Base Pairs: 1728bp
Origin: Sythetic
Properties: A coding sequence of xynA and PKC-OP.
Usage and Biology
BBa_K4096006 is a coding sequence of alkali cellulase (PKC-OP) from Pseudomonas aeruginosa and xylanase (xynA) from Bacillus subtilis. Cellulases break down the cellulose molecule into monosaccharides ("simple sugars") such as beta-glucose, or shorter polysaccharides and oligosaccharides. Several different kinds of cellulases are known, which differ structurally and mechanistically. Xylanase and β-xylosidase are the backbone degrading enzymes of heterogeneous xylan, which can degrade the xylan backbone to produce xylo-oligosaccharides or xyloses with different degrees of polymerization, which play an important role in the degradation of hemicellulose.
Construct design
Xylanase has the properties of hydrolyzing hemicellulose and can cooperate with cellulase to promote the biotransformation of lignocellulose. Add the Bacillus subtilis xylanase xynA gene to the cloning recombinant vector to make it compatible with cellulase PKC-OP and express β-xylosidase at the same time (as shown in the figure below).
The new family β-xylosidase from Humicola insolens Y1 has high tolerance to D-xylose. The gene is linked to a vector expressing xylanase xynA through GS linker to form pSIP403-PUS-xynA -xyl3A recombinant plasmid.
The profiles of every basic part are as follows:
BBa_K4093000
Name: xynA
Base Pairs: 642bp
Origin: Bacillus subtilis
Properties: endo-1,4-beta-xylanase
Usage and Biology
BBa_K4093000 is a coding sequence of from Bacillus subtilis. Xylanase is a kind of complex enzyme preparation specialized in degrading xylan in cereals.
BBa_K4096002
Name: PKC-OP
Base Pairs: 1086bp
Origin: Pseudomonas aeruginosa, genome
Properties: A coding sequence of alkali cellulase.
Usage and Biology
BBa_K4096002 is a coding sequence of alkali cellulase (PKC-OP) from Pseudomonas aeruginosa. Cellulases break down the cellulose molecule into monosaccharides ("simple sugars") such as beta-glucose, or shorter polysaccharides and oligosaccharides.
Experimental approach
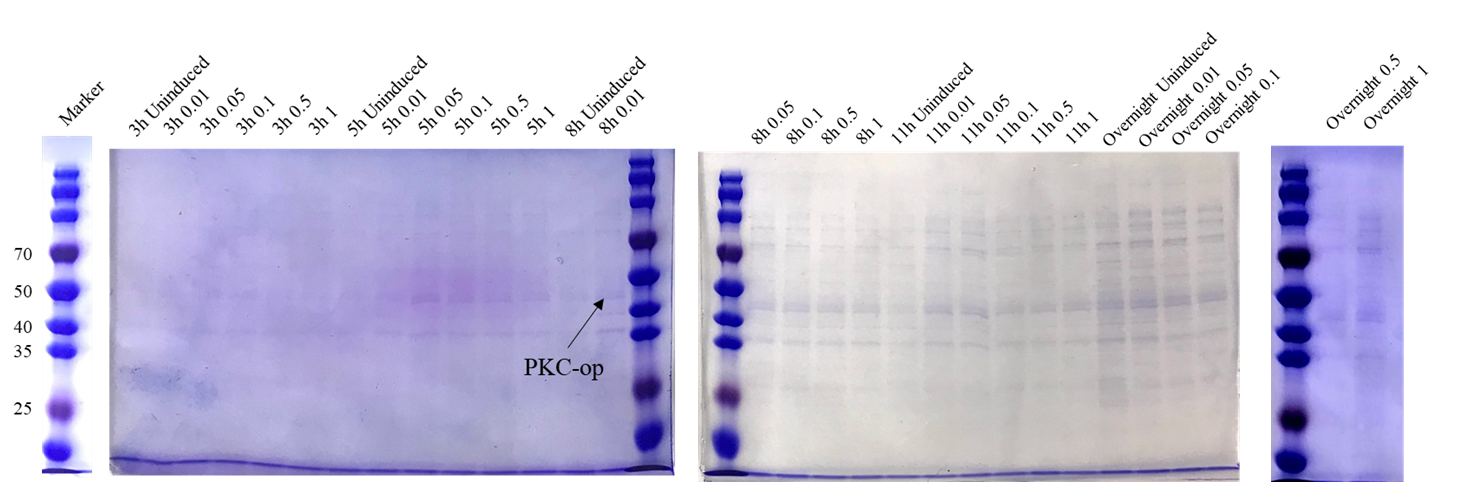
As the pET-25b vector contains a pelB signal peptide, the engineered strain would secret the protein into the medium. Therefore, we collected the culture supernatant after induction and ran SDS-PAGE for verification (Figure 3).
The theoretical molecular weight of the PKC cellulase is 45.6 kDa. As seen from the SDS-PAGE (Figure 3), there is a wide band just above 40 kDa and it indicates that we have obtained the PKC cellulase.
For xynA, we put it to SDS-PAGE (SDS-polyacrylamide gel electrophoresis) which could determine the level of protein expression. As seen in the gel map (Maker, culture solution, intracellular supernatant, intracellular precipitation), the second red line of Marker represents 25KDa and the target protein should be 23.28kDa. The blue line in the culture solution was near 23 KDa, it indicates that our protein could be successfully expressed in E. coli. We will further add the Bacillus subtilis xylanase xynA gene to the cloning recombinant vector to make it compatible with cellulase PKC-OP and express β-xylosidase at the same time. We wish that in this way, we will be able to improve the utilization rate of silage and grain mixed feed, and the applicable varieties of poultry and ruminants.
References
1. MUCK R,NADEAU E,MCALLISTER T,et al. Silage review: recent advances and future uses of silage additives [J]. J Dairy Sci
2. 刘海燕,张鹏举,王秀飞,等. 正交试验法优化玉米秸秆穰叶青贮发酵剂的研究 [J]. 中国饲料,2018 ( 11) : 74-79.
3. Coward-Kelly G., Aiello-Mazzari C., Kim S., Granda C., and Holtzapple M., 2003, Suggested improvements to the standard filter paper assay used to measure cellulase activity,Biotechnology & Bioengineering, 82(6): 745-749.
4. Ghose T.K., 1987, Measurement of cellulase activities, Pure & Appl Chem, 59(2): 257-268.
5. Lu Z.L., Chen D., Zhang S.S., Lu Q., and Huang R.B., 2012, A Pseudomonas aeruginosa producing alkaline cellulase, China Patent, 2012103821716
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 1723
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |