Part:BBa_K4000005
TEF1p+GA+CYC1t
Profile
Name: TEF1p+GA+CYC1t
Base Pairs: 2251 bp
Origin: Saccharomyces cerevisiae, Saccharomycopsis fibuligera, synthesis
Properties: An expression box of GA
Usage and Biology
Alcohol (ethanol) is an important chemical in the fields of food, medicine and bio-fuel. In particular, since the outbreak of COVID-19, the demand for alcohol for disinfection has skyrocketed [1]. Liquefaction enzyme expression in Saccharomyces cerevisiae has been reported. Bacillus amyloliquefaciens, Bacillus stearothermophilus, Bacillus Licheniformis, Bacillus subtilis Subtillis, Streptococcus Bovis, Debaryomyces Occidentali, Saccharomycopsis fibuligera and barley are common sources of α -amylase [2-3]. Because that the yeast must secrete enough amylase to support its growth at a higher initial starch concentration, and only growth can secrete enough amylase. This paradox leads to a prolonged yeast fermentation cycle, which cannot meet the requirements of industry.
Based on the above studies [4-5], we considered codon optimized Glucoamylase gene (GA) derived from S. fibuligera. After elements optimization, the saccharification enzyme activity of culture supernatant of the strain obtained and the alcohol production capacity of fermented corn starch subenzyme were tested in order to construct the proper strain.
Construct design
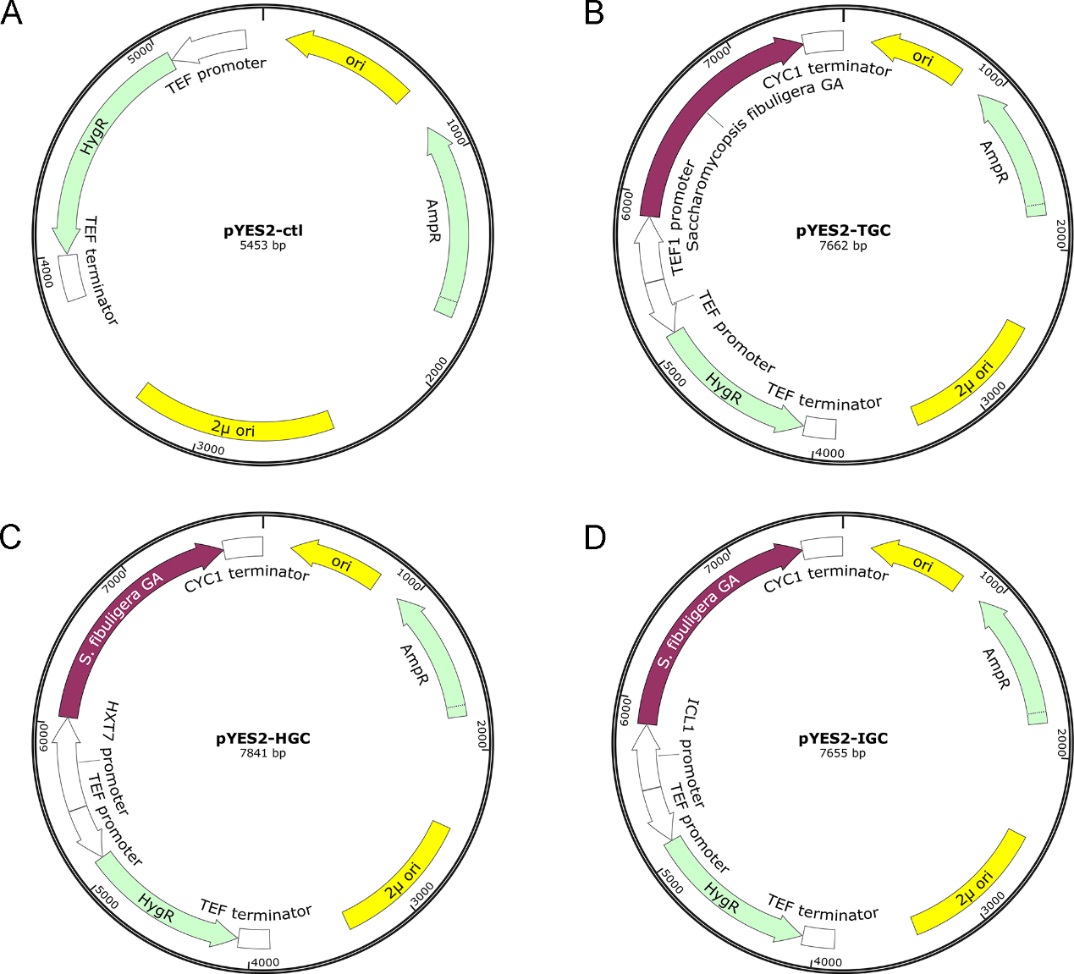
GA (Glucoamylase) is known as the type of enzyme that can easily break down starches into glucose, which afterwards becomes usable and absorbable. To optimize the reaction system, we choose three different promoters: the constitutive promoter TEF1, the glucose-inducible promoter HXT7, and the glucose-repressive promoter ICL1. The transcription of GA gene also controlled by strong terminator CYC1(Figure 2).
The profiles of every basic part are as follows:
BBa_K4000000
Name: TEF1p
Base Pairs: 434bp
Origin: Saccharomyces cerevisiae, genome
Properties: A constitutive promoter
Usage and Biology
TEF1 gene have a strong promoter activity, and it has been shown to be constitutively expressed even in the presence of glucose. The TEF1 promoter could be used for production of homologous or heterologous proteins under conditions where the expression of a large number of genes involved in the use of less favoured carbon sources are repressed.
BBa_K40000001
Name: GA
Base Pairs: 1567bp
Origin: Saccharomycopsis fibuligera, synthesis
Properties: An enzyme that can easily break down starches into glucose
Usage and Biology
Glucoamylase is an enzyme that can be obtained from the yeast or fungi in the Aspergillus genus such as Aspergillus niger. The enzyme decomposes starch molecules in the human body into the useful energy compound of glucose. This is accomplished by removing the alpha-1 and 4-glycosidic linkages from the non-reducing end of the starch molecule. These molecules are more commonly referred to as polysaccharides and are frequently either amylase- or amylopectin-based.
BBa_K4000002
Name: CYC1t
Base Pairs: 250bp
Origin: Saccharomyces cerevisiae, genome
Properties: Common transcriptional terminator
Usage and Biology
This is a common transcriptional terminator. Placed after a gene, it completing the transcription process and impacting mRNA half-life. This terminator can be used for in vivo systems,and can be used for modulating gene expression in yeast.
BBa_K4000003
Name: HXT7p
Base Pairs: 587bp
Origin: Saccharomyces cerevisiae, genome
Properties: Inducible promoter regulated by glucose
Usage and Biology
HXT7 promoter is one of the high-affinity hexose transporter, it is sufficient for complementary expression of invertase to restore the growth of Saccharomyces cerevisiae in raffinose medium. HXT7 promoter could be used for heterologous protein expression in S. cerevisiae.
BBa_K4000004
Name: ICL1p
Base Pairs: 401bp
Origin: Saccharomyces cerevisiae, genome
Properties: Inducible promoter regulated by glucose
Usage and Biology
The ICL1 promoter displays high levels of induced expression; however, ICL1 repression was incomplete in the presence of glucose.
Experimental approach
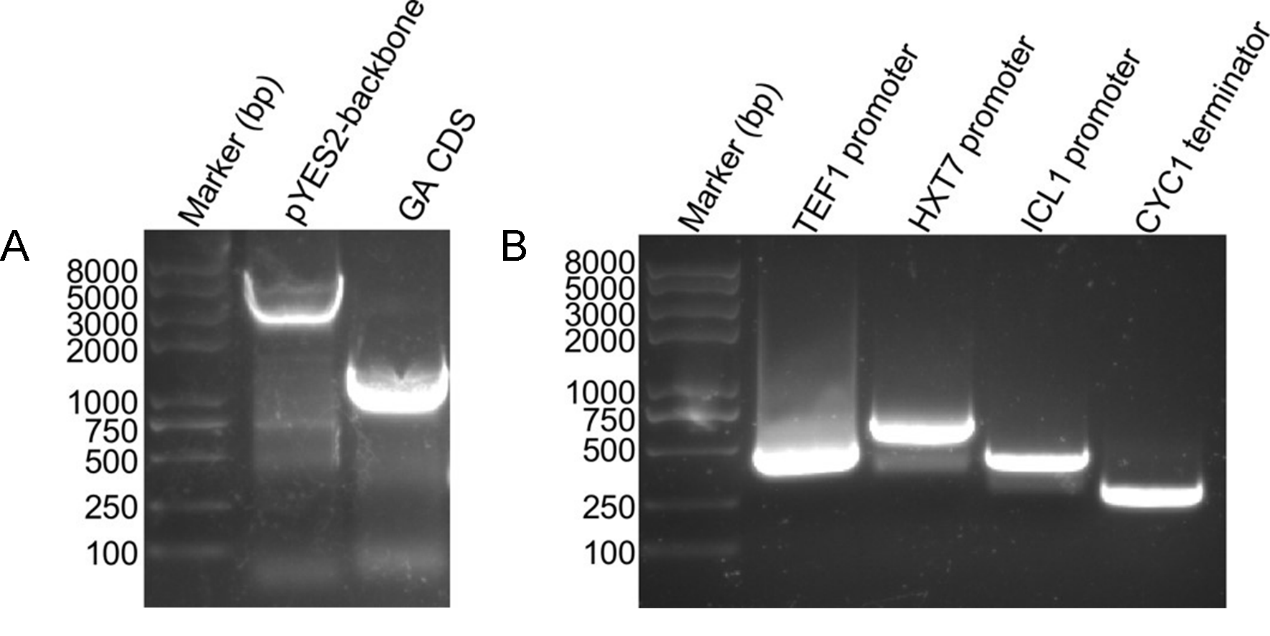
1. Fragments PCR products Electrophoresis
The basic parts of the plasmids such as the pYES2 backbone, GA coding sequence, 3 promoters, and terminators were all amplified successfully firstly (Fig. 3 A and B). Then the multiplex GA expression cassettes were assembled using the overlap PCR method.
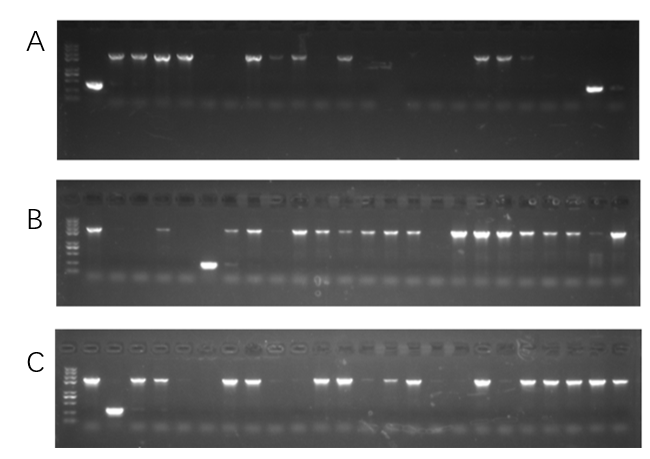
2. Colony PCR to identify the correct plasmids
We chose 24 colonies to verify whether the plasmids were correct or not using the colony PCR, the positive rate for the plasmids pYES2-TGC(Figure 4A), pYES2-HGC(Figure 4B), and pYES2-IGC(Figure 4C) were 11/24, 17/24, and 17/24, respectively. Two or three positive colonies were sequenced to verify further.
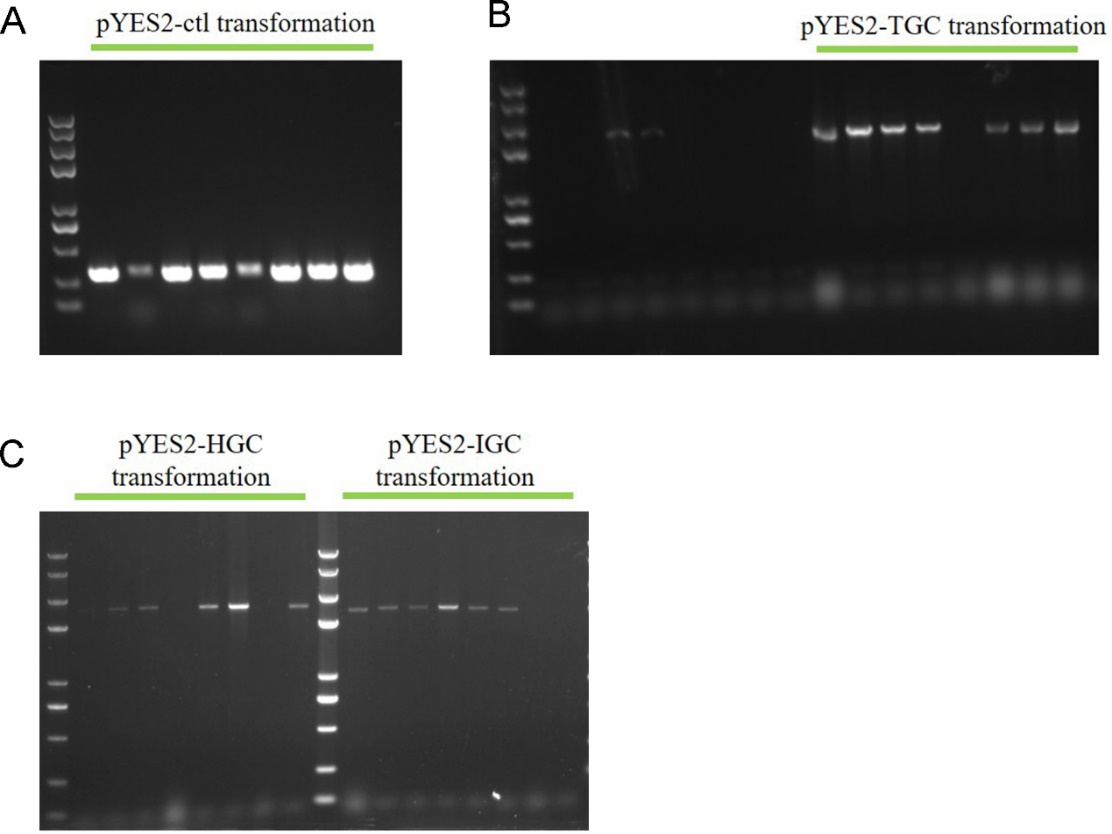
3. GA plasmids transformation
GA-expressing plasmids transformation and positive S. cerevisiae transformants selection using 50 μg/mL (Figure 5A) and 350 μg/mL (Figure 5B) hygromycin. Top left, pYES2-ctl. Top right, pYES2-TGC. Bottom left, pYES2-HGC. Bottom right, pYES2-IGC.
4. Colony PCR to identify the correct transpormed S. cerevisiae
The colonies which grew on the high concentration hygromycin plates were subjected to colony PCR to verify the plasmids transformation again. From Fig. 6 we can see that positive bands implied the plasmids transformation successfully.
Proof of function
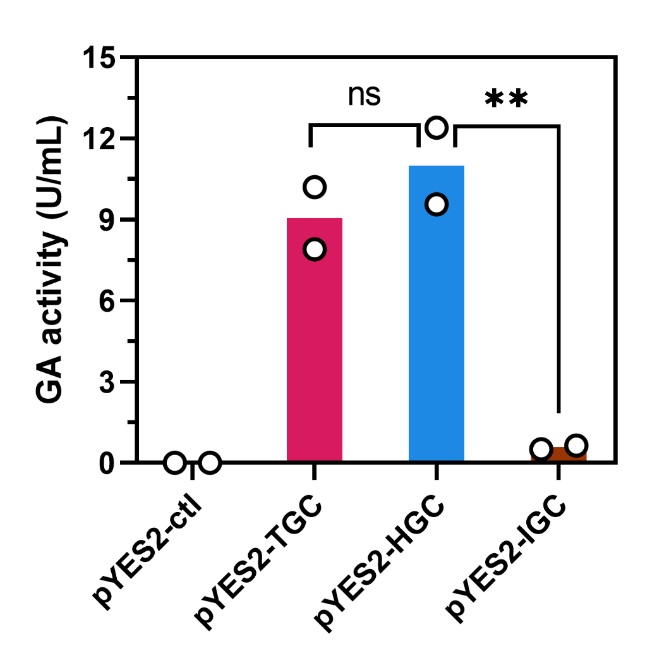
1. Glucoamylase activity assay
S. cerevisiae strains harboring various plasmids glucoamylase activity determination. **Statistical significance between indicated strains by Student’s t test, p < 0.01. ns, not significant. Data represent the means of two independent colonies.
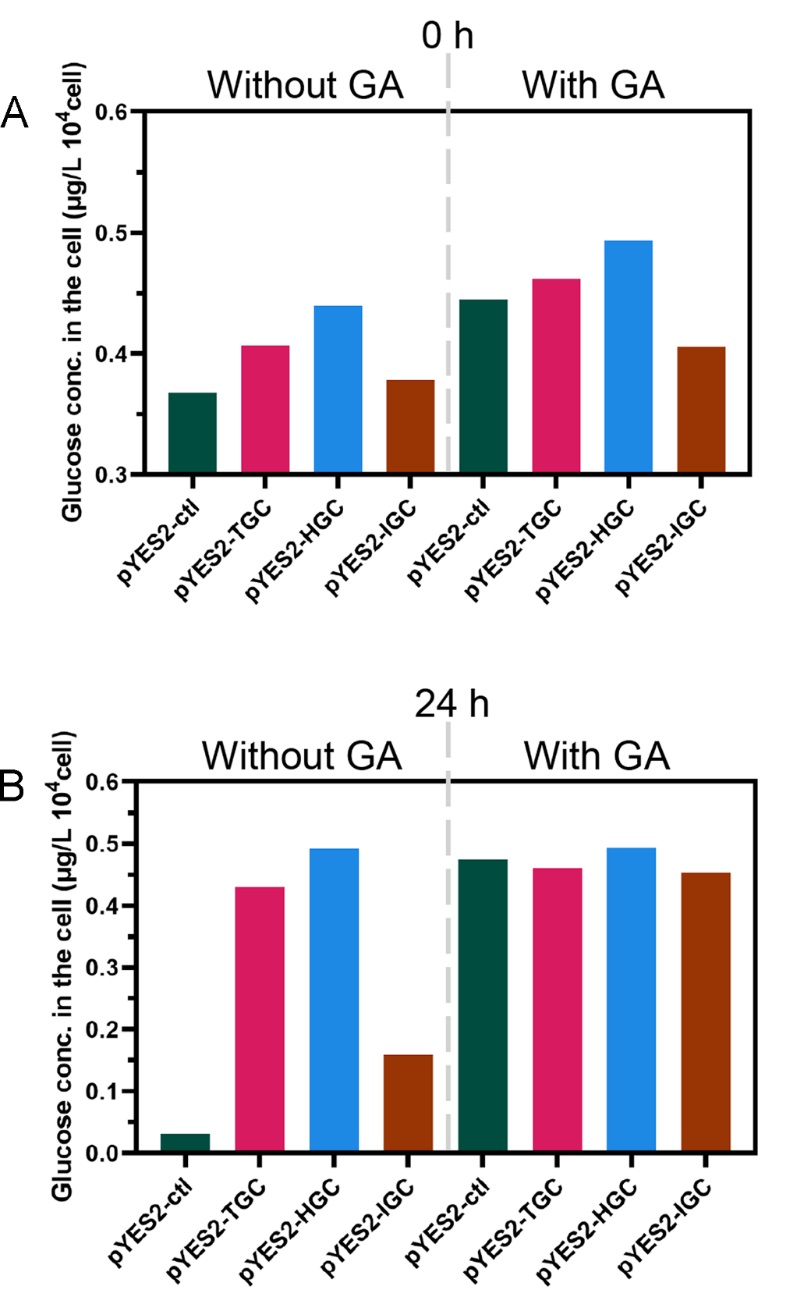
2. Fermentation test
To verify the GA secretion capacity of the GA-expressing S. cerevisiae strains, we measured the glucose concentration inside the cell during the corn starch fermentation process. As shown in Fig. 6A, at the initial stage (0 h), when the GA was added during the “starch-to-glucose” process, higher contents of glucose were detected than the process without GA addition.
Improvement of an existing part
Compared to the old part BBa_K801081, a TEF1 promoter, the old part BBa_K2391000, a ICL1 promoter, and the old part BBa_K3930019, we design a new part BBa_K4000005, which contains the three old promoters and a new protein GA. The GA protein is a (Glucoamylase) is known as the type of enzyme that can easily break down starches into glucose.
The group iGEM21_Toulouse_INSA-UPS aimed to characterize the rtTA promoter composed of HXT7 promoter to produce β-carotene. They also usde TetO7 promoter and found that promoter TetO7 is leaky, but showed less results of rtTA promoter. So as to the old part BBa_K801081 and BBa_K2391000, they showed little information of those promoters.
Based on the these groups’ contribution, our team design the new composite part BBa_K4000005 to express GA regulated by different promoters. After the composite part was inserted in a particular plasmid vector and transformed into S. cerevisiae. The different regulatory properties in the fermentation are measured, so as to achieve the purpose of our project.
First of all, we constructed a composite part BBa_K4000005 and transformed it into S. cerevisiae. We successfully tested the property during the starch to glucose process.
Furthermore, in order to have a general idea of the best promoter, we set up different time knots to measure the glucose concentration inside the cell during the corn starch fermentation. The experimental data shows that the optimal promoter is TEF1. In addition, our project can be used in industrial biofactory companies. The recommended particularly components could be used for the application of their patented-strain protection technology. Therefore, our engineering strain has a huge potential commercial market.
References
1. Görgens J F, Bressler D C, van Rensburg E. Engineering Saccharomyces cerevisiae for direct conversion of raw, uncooked or granular starch to ethanol[J]. Critical reviews in biotechnology, 2015, 35(3): 369-391.
2. Van Zyl W H, Bloom M, Viktor M J. Engineering yeasts for raw starch conversion[J]. Applied microbiology and biotechnology, 2012, 95(6): 1377-1388.
3. Maury J, Kannan S, Jensen N B, et al. Glucose-dependent promoters for dynamic regulation of metabolic pathways[J]. Frontiers in bioengineering and biotechnology, 2018, 6: 63.
4. Weber E, Engler C, Gruetzner R, et al. A modular cloning system for standardized assembly of multigene constructs[J]. PloS one, 2011, 6(2): e16765.
5. Pollak B, Cerda A, Delmans M, et al. Loop assembly: a simple and open system for recursive fabrication of DNA circuits[J]. New Phytologist, 2019, 222(1): 628-640.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 565
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 526
Illegal AgeI site found at 1897 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 189
Illegal SapI site found at 3
Illegal SapI.rc site found at 426
| None |