Part:BBa_K4000001
GA
Improvement by Team iGEM23_SubCat-China
BBa_K4000001 (GA)
Group: SubCat-China iGEM 2023
Summary
We constructed a new recombinant plasmid BBa_K4845020 (X-2-GA-2) based on the old part BBa_K4000001 (GA). X-2-GA-2 was transferred into yeast 1974 for protein expression, enzyme activity detection, quantitative and qualitative detection of its ability to decompose starch. It was proved by iodine staining experiment that X-2-GA-2 could decompose starch, and the ability of recombinant Saccharomyces cerevisiae to decompose starch was further characterized by measuring alcohol production.
Compared with the old part BBa_K4000001 (GA), the new recombinant plasmid BBa_K4845020 (X-2-GA-2) has two main improvements. First, we added two GAs to the backbone X-2 plasmid to improve the ability to decompose starch. At the same time, the optimal expression conditions of the protein GA were explored and added to the test experiments. The starch degradation ability of recombinant X-2-GA-2 in Saccharomyces cerevisiae was characterized by quantitative and qualitative detection methods. Secondly, on the basis of adding α-amylase, glucoamylase was further integrated into the chromosome of S. cerevisiae. Based on the hydrolysis circle, the starch degradation ability of the above strains was tested. The hydrolase activity of the fermentation supernatant of the obtained strains was tested, and the ability of the strains to ferment raw starch of sweet potato residue to produce alcohol was tested. Finally, S. cerevisiae strains with autocrine α-amylase and glucoamylase were obtained to achieve the goal of saving enzyme dosage. The results showed that the alcohol production capacity of the S. cerevisiae strains with autocrine α-amylase and glucoamylase was indeed higher than that of iGEM21_Fujian_United.
Construction Design and Engineering Principle
The problems we are going to solve are that the cost of exogenous enzymes during the alcohol fermentation process is much too high, and wasted sweet potato residue is harmful to the environment[1-3]. Therefore, as long as we enable the Saccharomyces cerevisiae to self-secrete alpha-amylase and glucoamylase, which function to completely hydrolyze starch into glucose molecules through synthetic biology, then we can largely reduce the cost of exogenous enzymes, and we can also put sweet potato residue into use as a raw material of alcoholic fermentation to turn the pollution problems into profits and efficiency smartly. To be more specific, we will transform plasmids BBa_K4845020 (X-2-GA-2) containing genes that express alpha-amylase and glucoamylase into our yeast 1974 (Figure 1)[4-5]. Therefore, we achieve reducing the cost of addition of exogenous enzymes during alcoholic fermentation.

Figure 1: Overview of the methodology of our project design through synthetic biology (made in Canva.cn)
Characterization/Measurement
A) Method of Transparent Circle
According to the property of starch that turns blue as it meets iodine solution, we placed our constructed Saccharomyces cerevisiae in the culture dish with starch solution distributed evenly. If our saccharomyces cerevisiae is successfully constructed, there will be alcohol produced around the strain because of our engineered property of self-secreting amylase and glucoamylase which work to decompose starch into glucose molecules, and those glucose molecules will be fermented by our constructed yeast cells 1974. As shown in Figure 2A, B, C, our constructed yeast cell did function to turn starch into alcohol, giving the phenomenon that there are transparent circles with respectively diameters of 2.14cm, 2.56cm, and 2.23cm around our engineered yeast cell.

Figure 2: Transparent circle experiment for the function testing
Diameter of the transparent circle in A: 2.14cm
Diameter of the transparent circle in B: 2.56cm
Diameter of the transparent circle in C: 2.23cm
B) Enzyme Activity Detection in Starch Hydrolyzing Capacity
To verify the GA and temA activity, we measured the enzyme activity of the recombinase. The enzyme activity was measured by the glucose content detection kit. Enzyme activity was expressed as U/mL supernatant, and one unit of enzyme activity was defined as the amount of enzyme required to release 1 μmol glucose per minute. The recombinant was incubated at different pH values (3, 4, 5, 6, and 7) and temperature values (30℃, 40℃, 50℃, 60℃, 70℃, and 80℃) to study the enzymatic properties of the recombinant enzyme (Figure 3).

Figure 3: The enzyme activity of Yeast 1974-GA-temA and Wild Yeast 1974 under different pH value at the same temperature
According to Figure 3A, we can see that generally, Yeast 1974-GA-temA possesses higher enzyme activity than the wild at 30 °C. In addition, we can tell that when the pH value equals 5, both strains reach their highest enzyme activity of both strains where the temperature is lower than 50 °C, and there seem to be little changes in the enzyme activity responding to the pH values after the temperature equals or is higher than 50 °C.

Figure 4: The enzyme activity of Yeast 1974-GA-temA and Wild Yeast 1974 under different temperature at the same pH value
According to Figure 4, when the temperature goes higher, the enzyme activity of both strains decreases basically. Based on the curve trends of Figure 4-A, B, C, E, there is an obvious turning point at 50 °C, which we have already pointed out previously. But it is worth noting that when the pH value equals 5, both strains show a different trend of enzyme activity against temperature, and it will require further research. Compared with the wild, Yeast 1974-GA-temA possesses higher enzyme activity when the pH value is higher than 5.

Figure 5: The comparison of the enzyme activity curves of Yeast 1974-GA-temA under different pH values and different temperature, respectively
After we integrated Figure 3 and Figure 4, we can obtain the comparison graphs in Figure 5. To conclude, the enzyme activity of the recombinant enzyme in Yeast 1974-GA-temA is highest at pH 5 and 30 °C at the given setting. Also based on our data, the proper condition for our recombinant enzyme will be when the pH value range is 4 to 6 and the temperature is 30 °C around where our recombinant yeast possesses better enzyme activity in starch hydrolyzing capacity than the wild.
C) Determination of alcohol production
To further and directly verify the normal functioning of our constructed yeast, we applied them to produce alcohol in reality, thus obtained this bar chart. Figure shows that the 1974-GA-temA did produce incremental alcohol over time, and it does boost the alcohol production a lot in comparison to the control group yeast 1974 (Figure 6). The alcohol production capacity of 2022 iGEM21_Fujian_United was 0.18g / L. The alcohol production of our combined yeast 1974-GA-temA was 0.23 g / L, which proved that the alcohol production capacity of our combined yeast 1974-GA-temA was improved.

Figure 6: Alcohol production of yeast 1974 and GA-temA-1974
Reference
[1] Bušić Arijana, Marđetko Nenad, Kundas Semjon, et al. Bioethanol Production from Renewable Raw Materials and Its Separation and Purification: A Review[J]. Food technology and biotechnology, 2018, 56(3): 289-311.
[2] Xu Shuai, Yang Li, Tan Liping, et al. Enzymatic hydrolysis and application of sweet potato residue [ J ]. Journal of Qilu University of Technology. 2021, 35(03): 28-33.
[3] Xin Wang, Bei Liao, Zhijun Li, et al. Reducing glucoamylase usage for commercial-scale ethanol production from starch using glucoamylase expressing Saccharomyces cerevisiae[J]. Bioresources and Bioprocessing, 2021, 8(1): 20.
[4] Rosemary A. Cripwell, Lorenzo Favaro, Marinda Viljoen-Bloom, et al. Consolidated bioprocessing of raw starch to ethanol by Saccharomyces cerevisiae: Achievements and challenges[J]. Biotechnology Advances, 2020, 42: 107579.
[5] LM de Moraes, S Astolfi-Filho, SG Oliver. Development of yeast strains for the efficient utilization of starch: evaluation of constructs that express alpha-amylase and glucoamylase separately or as bifunctional fusion proteins[J]. Applied microbiology and biotechnology, 1995, 43(6): 1067-76.
Profile
Name: GA
Base Pairs: 1567bp
Origin: Saccharomycopsis fibuligera, synthesis
Properties: An enzyme that can easily break down starches into glucose
Usage and Biology
Glucoamylase is an enzyme that can be obtained from the yeast or fungi in the Aspergillus genus such as Aspergillus niger. The enzyme decomposes starch molecules in the human body into the useful energy compound of glucose. This is accomplished by removing the alpha-1 and 4-glycosidic linkages from the non-reducing end of the starch molecule. These molecules are more commonly referred to as polysaccharides and are frequently either amylase- or amylopectin-based.
Experimental approach
1.GA plasmids transformation
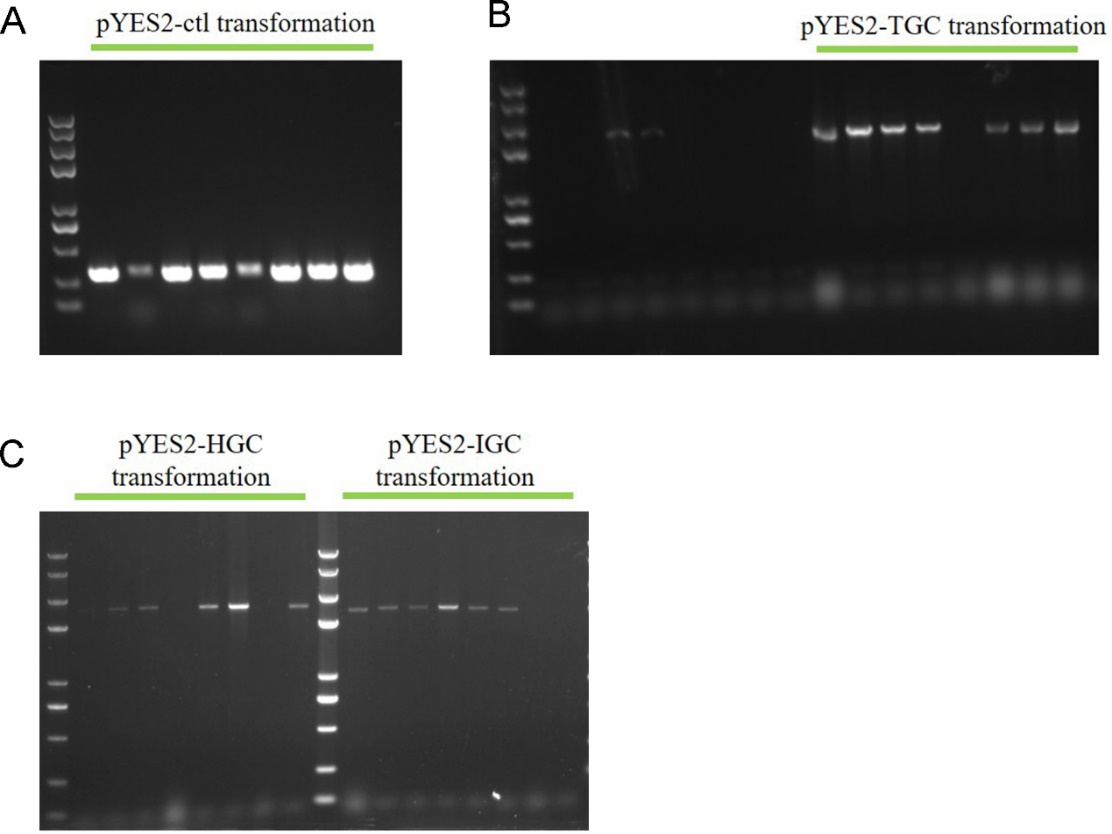
GA-expressing plasmids transformation and positive S. cerevisiae transformants selection using 50 μg/mL (Figure 2A) and 350 μg/mL (Figure 2B) hygromycin. Top left, pYES2-ctl. Top right, pYES2-TGC. Bottom left, pYES2-HGC. Bottom right, pYES2-IGC.
2. Colony PCR to identify the correct transpormed S. cerevisiae
The colonies which grew on the high concentration hygromycin plates were subjected to colony PCR to verify the plasmids transformation again. From Fig. 3 we can see that positive bands implied the plasmids transformation successfully.
Proof of function
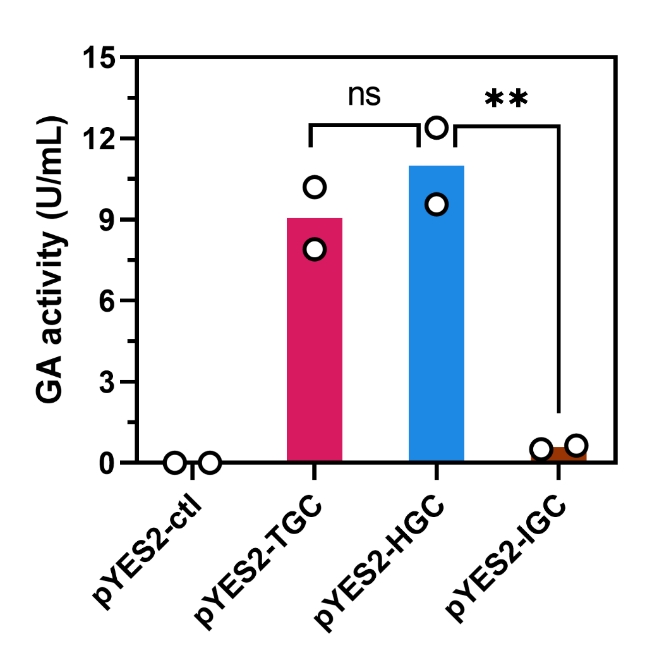
1. Glucoamylase activity assay
S. cerevisiae strains harboring various plasmids glucoamylase activity determination. **Statistical significance between indicated strains by Student’s t test, p < 0.01. ns, not significant. Data represent the means of two independent colonies.
2. Fermentation test
To verify the GA secretion capacity of the GA-expressing S. cerevisiae strains, we measured the glucose concentration inside the cell during the corn starch fermentation process. At the initial stage (0 h), when the GA was added during the “starch-to-glucose” process, higher contents of glucose were detected than the process without GA addition.
References
1. Görgens J F, Bressler D C, van Rensburg E. Engineering Saccharomyces cerevisiae for direct conversion of raw, uncooked or granular starch to ethanol[J]. Critical reviews in biotechnology, 2015, 35(3): 369-391.
2. Van Zyl W H, Bloom M, Viktor M J. Engineering yeasts for raw starch conversion[J]. Applied microbiology and biotechnology, 2012, 95(6): 1377-1388.
3. Maury J, Kannan S, Jensen N B, et al. Glucose-dependent promoters for dynamic regulation of metabolic pathways[J]. Frontiers in bioengineering and biotechnology, 2018, 6: 63.
4. Weber E, Engler C, Gruetzner R, et al. A modular cloning system for standardized assembly of multigene constructs[J]. PloS one, 2011, 6(2): e16765.
5. Pollak B, Cerda A, Delmans M, et al. Loop assembly: a simple and open system for recursive fabrication of DNA circuits[J]. New Phytologist, 2019, 222(1): 628-640.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 131
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 92
Illegal AgeI site found at 1463 - 1000COMPATIBLE WITH RFC[1000]
| None |