Part:BBa_K3921006
dicF
DicF is a 53-nucleotide RNA molecule that can inhibit the translation process of ftsZ mRNA.
FtsZ is closely related to the cell division of bacteria. Escherichia coli without FtsZ will not be able to form a division septum, causing cell to become filamentous [1]. DicF is a 53-nucleotide RNA molecule that can inhibit the translation process of ftsZ mRNA [2]. In 2017, Lin-Chao et al explored the mechanism of DicF under anaerobic conditions and found that the expression of DicF led to the filamentous morphology of MG1655. They ectopically expressed DicF in MG1655, and filamentous cells were observed under aerobic conditions [3]. We cloned the gene dicF into pBAD24 plasmid to obtain pBAD-dicF plasmid, so that sRNA DicF can be induced by arabinose promoter. After transforming pBAD-dicF into MG1655, when the culture medium OD600 was 0.3, it was induced with arabinose (0.2%) for 12 hours and then observed under microscope. As shown in Fig. 1A, compared with the strain pBAD-dicF MG1655 without arabinose (Fig. 1B), most of the strains induced by arabinose were filamentous, indicating that the process of cell division was inhibited.We calculated the relative length of MG1655 cells in terms of the pixels of the microscope photos, and the relative length of cells induced with L-arabinose increased by about 3 times (Fig. 2).
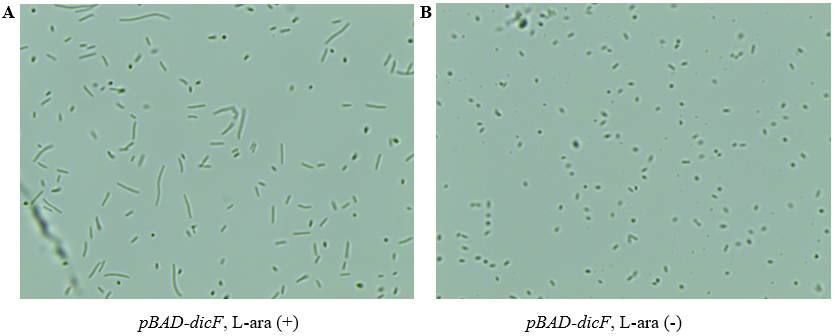
Figure 1. Microscopic observation map of pBAD-dicF MG1655. (A) Induced by adding L-arabinose. (B) Do not add L-arabinose. (A) and (B) were cultured in 200rpm shaker at 37 ℃ for the same time.

Figure 2. Statistical diagram of MG1655 cell length (In pixels).
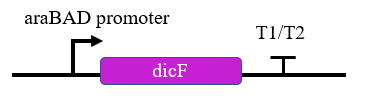
Figure 3. Schematics of the dicF circuit.
References
[1] Weiss, D. S. Bacterial cell division and the septal ring. Mol. Microbiol. 54, 588–597 (2004).
[2] Tétart, F. & Bouché, J. ‐P. Regulation of the expression of the cell‐cycle gene ftsZ by DicF antisense RNA. Division does not require a fixed number of FtsZ molecules. Mol. Microbiol. 6, 615–620 (1992).
[3] Murashko, O. N. & Lin-Chao, S. Escherichia coli responds to environmental changes using enolasic degradosomes and stabilized DicF sRNA to alter cellular morphology. Proc. Natl. Acad. Sci. U. S. A. 114, E8025–E8034 (2017).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |
