Part:BBa_K3852005
T1R1
The protein encoded by this gene is a G protein-coupled receptor and is a component of the heterodimeric amino acid taste receptor T1R1+3. The T1R1+3 receptor responds to L-amino acids but not to D-enantiomers or other compounds. Most amino acids that are perceived as sweet activate T1R1+3, and this activation is strictly dependent on an intact T1R1+3 heterodimer. Multiple transcript variants encoding different isoforms have been found for this gene.
T1R1 is essentially a recognition receptor for the nitrogen signal in the body, and it may recognize the umami substance through its amino group.In our experiments, we used this gene to express umami taste receptors in Saccharomyces cerevisiae.
Usage and Biology
Putative taste receptor. TAS1R1/TAS1R3 responds to the umami taste stimulus (the taste of monosodium glutamate). Sequence differences within and between species can significantly influence the selectivity and specificity of taste responses.We acquire it from synthesis company.
Experiment
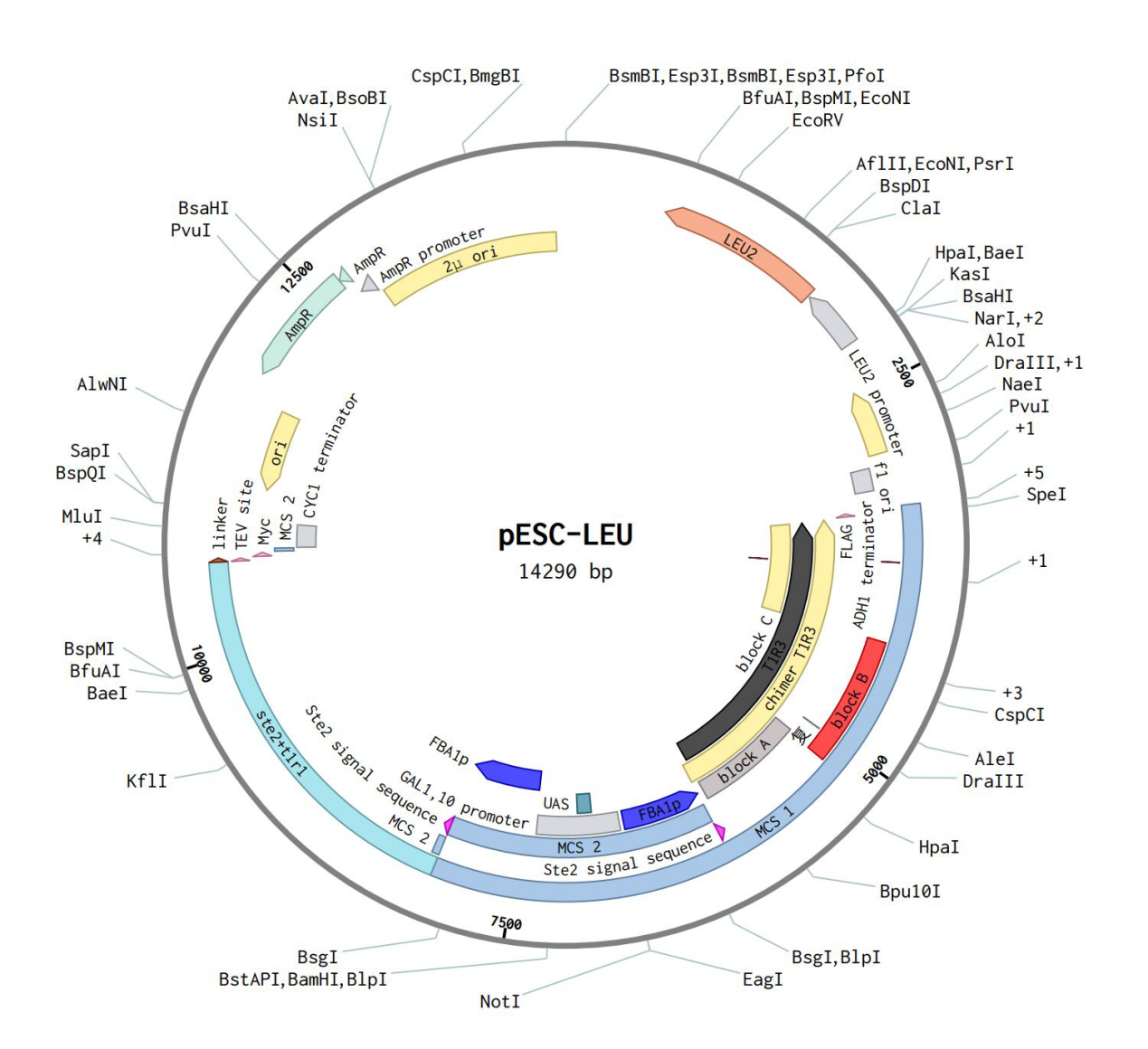
After literature research, we found that the taste receptors in the human body are made up of T1R1 and T1R3.So we plan to introduce T1R1 and T1R3 into the Saccharomyces cerevisiae. The first step is to integrate the two genes into pESC-LEU, and then use semi-lactose induction to allow the two genes to be expressed in the Saccharomyces cerevisiae at the same time.
The design of the plasmid is shown below:
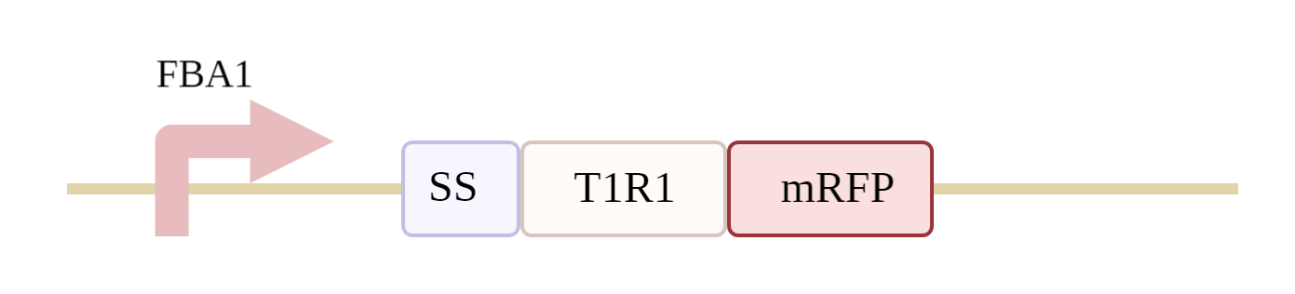
1. Modification and synthesis of T1R1 gene
We first obtain the sequence of T1R1 from NCBI, and then use Snap Gene for sequence optimization, which is mainly to modify the signal peptides of heterogenous receptors by replacing the first 20 Aa of the mGluR alpha of T1R1 (signal peptides of the protein) with Ste2,17 Aa, with the aim of increasing the content of T1R1 in the endoplasmic mesh, thereby increasing the chance of positioning it on the membrane. The gene synthesis company is then commissioned to optimize the cipher to synthesize the target gene we need.
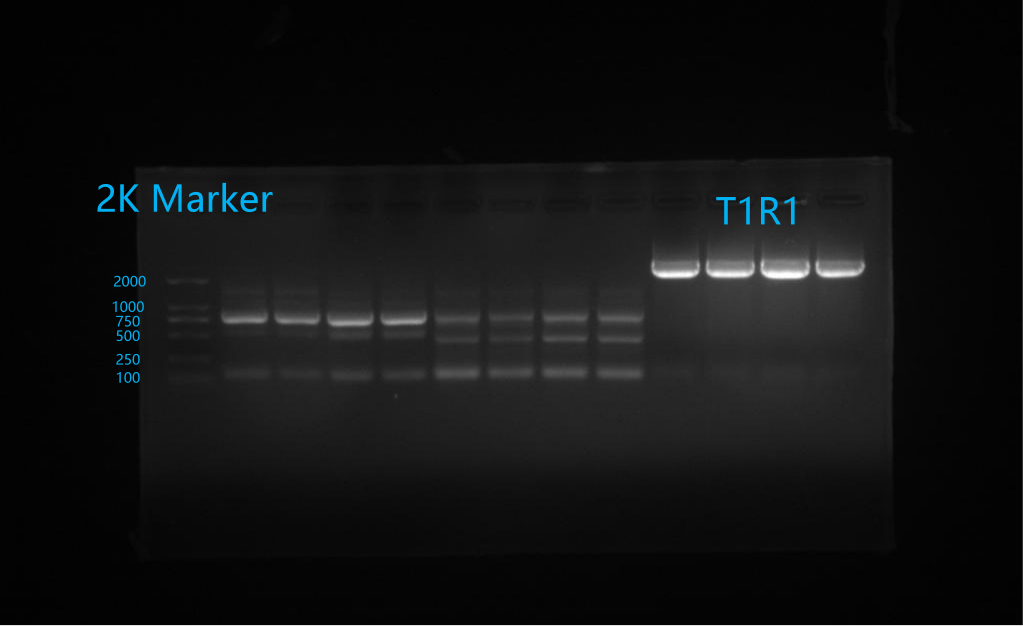
2. Agar sugar gel electrophoresis
Results: The genetic design of this purpose was successful, as shown in the following illustration.
3. E. coli conversion experiment
After successfully completing the construction of the synthetic T1R1 gene and plasmids, we introduced the receptor gene into E. coli by transformation experiment, cultured it with a culture medium added to ampicillin, and observed that colonies on the medium grew.
In order to verify the successful introduction of plasmids into E. coli, we carried out a bacterial liquid PCR, and then by running the verification glue, observed the correct strip, proving the successful construction of plasmids.
4. Electrical conversion of Saccharomyces cerevisiae
The plasmids extracted from E. coli were successfully imported into the Saccharomyces cerevisiae by electro-conversion.
5. The homologous recombination of Saccharomyces cerevisiae
After realizing the yeast electroconvert method to successfully introduce plasmids into the Saccharomyces cerevisiae, we then want to adopt a relatively simple new way to realize the gene introduction: the specific idea is to divide the binding site of YPRC3 into two parts and add to the left and right sides of the target gene, that is called the left and right homologous arm, and then transfer to the Saccharomyces cerevisiae, after which the yeast genome will be homologous recombination.
Reference
[1] Huang Yulin, Lu Dingqiang, Liu Hai et al. Preliminary research on the receptor-ligand recognition mechanism of umami by an hT1R1 biosensor[J] Food & Function.2019,10(3): 1280-1287.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 2491
Illegal XbaI site found at 15
Illegal XbaI site found at 854
Illegal XbaI site found at 944
Illegal XbaI site found at 1262 - 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 2491
- 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 2491
Illegal BglII site found at 218
Illegal BglII site found at 1820 - 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 2491
Illegal XbaI site found at 15
Illegal XbaI site found at 854
Illegal XbaI site found at 944
Illegal XbaI site found at 1262 - 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 2491
Illegal XbaI site found at 15
Illegal XbaI site found at 854
Illegal XbaI site found at 944
Illegal XbaI site found at 1262 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 1440
| None |