Part:BBa_K3743003
Destabilizing Domain (DD)
Part description
The destabilising domain (DD) of dihydrofolate reductase (DHFR), which has shown promise as a biologic tool and a prospective gene therapy approach, may be used to modulate protein abundance in vivo simply by administering its stabilising ligand, the commonly prescribed antibiotic trimethoprim (TMP). Dihydrofolate reductase is an eight-stranded central folded beta sheet that forms the main fold feature of the DHFR polypeptide backbone. Seven of these threads are parallel and the eighth is antiparallel. It converts dihydrofolate to tetrahydrofolate using NADPH as an electron donor, which can be converted to the types of tetrahydrofolate cofactors used in 1-carbon transfer chemistry and participates in the synthesis of purine and thymidylate. In humans, the DHFR enzyme is encoded by the DHFR gene. It is located in the q11 → q22 region of chromosome 5.
Usage
Dihydrofolate reductase (DHFR) destabilizing domain (DD) is used in our circuit to be fused with MS2 protein that acts as a riboswitch by its binding to small nuclear ribonucleoprotein. Destabilizing domain could be stabilizing by trimethoprim (TMP) which acts as a small molecule inhibitor. After stabilizing DD, MS2 will inhibit the circuit via binding to U2-nRNP.
Literature Characterization
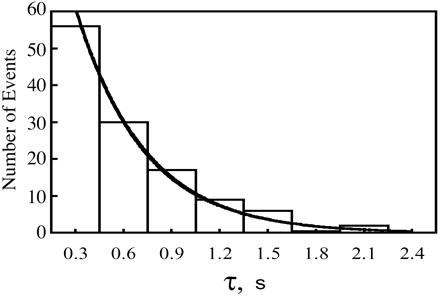
In one study that has done an exponential fit of the data as shown in figure (1). The study made a summary the ensemble averaged rate constants at various methotrexate concentrations with a limited degree of uncertainty due to the limited number of events and limited time resolution of the equipment.(1)
Characterization Of Mutational Landscape
A mutational landscape prediction through saturation mutagenesis of destabilizing domain (DD) and the effect of these mutations on the evolutionary fitness of the protein is tested after generating multiple sequence alignment of the protein sequence and predict mutational landscapes. As shown in the chart, the (L3N) mutation showed the highest score compared to other mutations. On the contrary, we can see that the (W73A) contributed to the lowest evolutionary fitness to DD. As shown in Figure (2)
Characterization by Mathematical Modelling
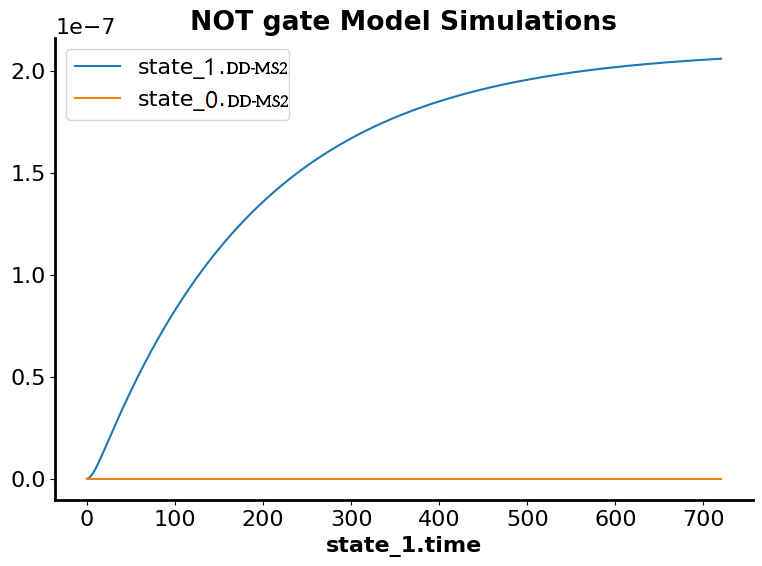
NOT Gate
In the simulation of mass action kinetics of DD_MS2 which is controlled by TMP, the blue plot represents that the steady state is achieved after 700 seconds in the absence of TMP and the red plot represent the inhibition of the transcription through binding of MS2 to small nuclear ribonucleoprotein (snRNP) which is controlled by TMP to terminate the transcription if the circuit is uncontrollable.showed increased transcription of the circuit which reach the steady state after about 200 seconds.
AND Gate
Blue plot achieves the maximum concentration at 1.6 after 700 seconds which represents the presence of 2 inputs in the AND (absence of TMP and binding of miRNA to toeholdOn switch upstream to the vaccine) and the red plot represents the inhibition of the transcription in absence of one of the two inputs wither the presence of the TMP so stabilized the destabilizing domain and inhibiting the circuit or absence of binding of miRNA to ToeholdOn switch
References
1.Rajagopalan, P. R., Zhang, Z., McCourt, L., Dwyer, M., Benkovic, S. J., & Hammes, G. G. (2002). Interaction of dihydrofolate reductase with methotrexate: ensemble and single-molecule kinetics. Proceedings of the National Academy of Sciences, 99(21), 13481-13486.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |