Part:BBa_K3743002
MS2
Part Description
This part produces a RNA bacteriophage Ms2 coat protein. The coat protein from the MS2 bacteriophage is a mem-ber of a group of small proteins that bind RNA in a multifunctional manner in related RNA bacteriophages.This protein is responsible for extending the life of the virus by binding to RNA stem-loop structure of 19 nucleotides which contain the initiation codon of the replicase gene it controls the sequence-specific RNA encapsidation and repression of replicase translation. Also it shuts off the synthesis of the viral replicase, switching the viral replication cycle to virion assembly instead of continued replication by binding the coat protein dimer to the hairpin
Usage
The final design of our vaccine include DD-MS2 riboswitch which represents an essential safety switch to control the transcriptions in extreme conditions by administering TMP that acts as a small molecule inhibitor which has the ability to stabilize the destabilizing domain (DD) that will free MS2 to be bound to its U2-small nuclear ribonucleoprotein that will inhibit the circuit to be controllable.
Literature Characterization
A study was conducted to induce mutation in the amino acid sequence of MS2 coat protein. Only those mutants which produced normal quantities of coat protein with the correct electrophoretic mobility were selected as shown in figure (1) to compare the differences regarding the stability and fitness.(1)
Characterization Of Mutational Landscape
A mutational landscape prediction through saturation mutagenesis of MS2 protein and the effect of these mutations on the evolutionary fitness of the protein is tested after generating multiple sequence alignment of the protein sequence and predict mutational landscapes. As shown in the chart, the (W70T) mutation showed the highest score compared to other mutations. On the contrary, we can see that the (L74G) contributed to the lowest evolutionary fitness to MS2. As shown in Figure (2)
Characterization by Mathematical Modelling
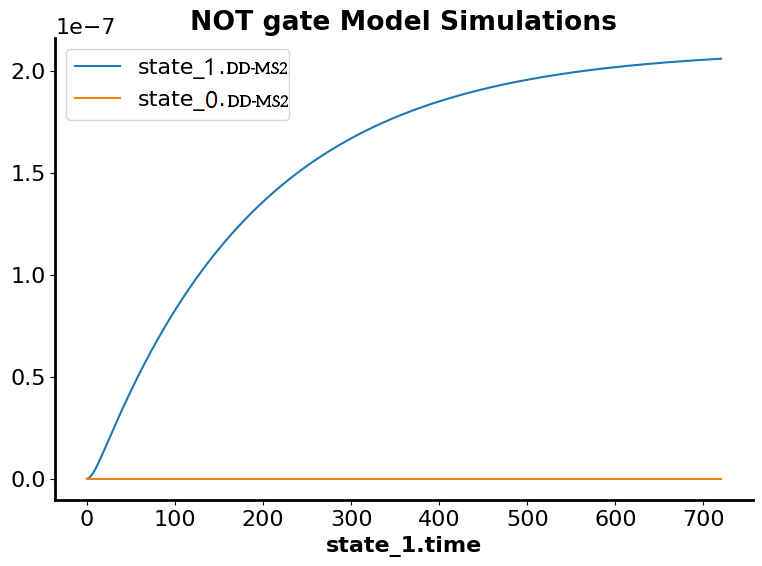
NOT Gate
In the simulation of mass action kinetics of DD_MS2 which is controlled by TMP, the blue plot represents that the steady state is achieved after 700 seconds in the absence of TMP and the red plot represent the inhibition of the transcription through binding of MS2 to small nuclear ribonucleoprotein (snRNP) which is controlled by TMP to terminate the transcription if the circuit is uncontrollable.showed increased transcription of the circuit which reach the steady state after about 200 seconds.
AND Gate
Blue plot achieves the maximum concentration at 1.6 after 700 seconds which represents the presence of 2 inputs in the AND (absence of TMP and binding of miRNA to toeholdOn switch upstream to the vaccine) and the red plot represents the inhibition of the transcription in absence of one of the two inputs wither the presence of the TMP so stabilized the destabilizing domain and inhibiting the circuit or absence of binding of miRNA to ToeholdOn switch
References
1.Peabody, D. S. (1993). The RNA binding site of bacteriophage MS2 coat protein. The EMBO journal, 12(2), 595-600.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 292
| None |