Part:BBa_K3407024
Fox-1 RBD* domain with depleted binding capacity, with T7 promoter - RBS - T1 terminator
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 55
Illegal BamHI site found at 444
Illegal XhoI site found at 453 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Contents
Usage and Biology
In many projects involving RNA approaches, the availability of a library of proteins able to bind strongly and specifically to an RNA target sequence represents a valuable and versatile set of BioBricks. Fox-1 RNA Binding Domain (RBD) is a protein domain able to recognise and bind a specific RNA sequence. Our designed part is mutated version of Fox-1 RBD (BBa_K3407004), where mutations F160A & F126A deplete its binding capacity [1] .
Fox-1 stands for Feminizing Locus on X, a protein acting as a numerator element to count the number of X chromosomes relative to ploidy. In these worms, Fox-1 is thought to determine sex by post-transcriptional repression of Xol-1. In mammalian genes, it strongly influences the splicing of alternative exons by binding to “UGCAUG” RNA sequence [1]. Orthologs are present in a wide range of multicellular eukarya species like zebrafish (Q642J5, Uniprot), mice (Q9JJ43, Uniprot), rats (A1A5R1, Uniprot), bulls (A6QPR6, Uniprot), frogs (Q66JB7, Uniprot), chicken (F1NCA4, Uniprot), polar bear (A0A384DA29, Uniprot), cats (A0A337SR52, Uniprot), platypuses (F6SR19, Uniprot), sunfishes (A0A3Q4B4Y5, Uniprot), tasmanian devils (G3VXH1 , Uniprot) and humans (Q9NWB1 , Uniprot).
-
 Figure 1: Crystal structure of Fox-1 RBD bound to its UGCAUGU target sequence. The structure has been resolved [1] available in PDB with 2ERR accession number 2ERR.
Figure 1: Crystal structure of Fox-1 RBD bound to its UGCAUGU target sequence. The structure has been resolved [1] available in PDB with 2ERR accession number 2ERR.
This protein has a domain of interaction with RNA called RNA Binding Domain (RBD), also commonly referred as RNA Recognition Motif (RRM) and Ribonucleoprotein (RNP) domain, referring to the domain formed by the residues 109 to 208 of Fox-1 (13.6 kDa when his-tagged). Its RBD has shown a high affinity (Kd = 0.49 nM at 150mM NaCl) to “UGCAUGU” RNA sequence where residues F126 and F160 play a crucial role in its binding capacity [1][2]. Interestingly, it has also shown a high specificity to the mentioned sequence where single mutations can reduce from 4,8 to more than 1.500 fold the affinity with its substrate [1], becoming a powerful BioBrick where a highly specific and affine interaction is needed between a protein and RNA. As shown in its crystal structure, C-terminal of the RBD is distant from the binding site and can accept a His-tag without showing a negative effect on the binding capacity [1], probably providing an adequate locus to fuse other domains or proteins. A mutated version of the protein domain has been expressed and tested as a control, where mutations F160A & F126A deplete its binding capacity [1] (BBa_K3407004).
Experimental results
Fox-1 RBD and RBD* (mutated) overexpression and purification
In order to check the overexpression of Fox-1 RBD and its mutated version (BBa_K3407004), we incubated the E. coli BL21 (DE3) (Negative control) and E. coli BL21 (DE3) transformed either with the pBbB7a_Fox-1_RBD (BBa_K3407020) or pBbB7a_Fox-1_RBD* (BBa_K3407024) at 37ºC until the OD600 reached ~ 0.6. We induced the cultures with a final IPTG concentration of 1 mM and incubated them 4h at 37ºC and overnight at 30ºC. The total protein content of the cells was analysed by SDS-PAGE electrophoresis (Figure 2A). As the recombinantly expressed Fox-1 RBD and RBD* (mutated) contain a His-tag at their N-terminal part, we purified them using affinity chromatography with the HisLink protein purification system (Promega). Samples obtained from the different purification steps were analysed by SDS-PAGE electrophoresis (Figure 2B).
-
 Figure 2: Purification of Fox-1 RBD and mutated Fox-1 RBD*. (A) SDS-PAGE gel of the overexpression samples before and after induction with IPTG. (B) SDS-PAGE of samples from the different purification steps. E. coli BL21 (DE3) is the negative control, E. coli BL21 (DE3) Fox-1 RBD contains the part BBa_K3407020 and E. coli BL21 (DE3) Fox-1 RBD* contains the part BBa_K3407024. MW (Molecular weight marker, #1610363 Bio-Rad), PI (pre-induction), 4h (4 hours after induction), ON (overnight), FT (flow-through), W1 and W2 (Washing), E (Elution). All the samples used corresponded to the same OD600.
Figure 2: Purification of Fox-1 RBD and mutated Fox-1 RBD*. (A) SDS-PAGE gel of the overexpression samples before and after induction with IPTG. (B) SDS-PAGE of samples from the different purification steps. E. coli BL21 (DE3) is the negative control, E. coli BL21 (DE3) Fox-1 RBD contains the part BBa_K3407020 and E. coli BL21 (DE3) Fox-1 RBD* contains the part BBa_K3407024. MW (Molecular weight marker, #1610363 Bio-Rad), PI (pre-induction), 4h (4 hours after induction), ON (overnight), FT (flow-through), W1 and W2 (Washing), E (Elution). All the samples used corresponded to the same OD600.
Additional protein bands corresponding to molecular weights of 13.6 kDa (Fox-1 RBD) and 13.4 kDa (Fox-1 RBD*, mutated) were observed after induction (Figure 2A), indicating that both proteins were overexpressed. Moreover, a clear band is obtained in the elution samples (E) (Figure 2B), demonstrating that both Fox-1 RBD and RBD* were successfully purified.
Fox-1 RBD* does not bind to its target sequence located in a shRNA loop
To study RNA-protein interaction, an Electrophoretic mobility shift assay (EMSA) was performed (Figure 3). When a protein binds to RNA, the complex migrates slower in the gel, shifting the RNA band to a higher position. shRNA (BBa_K3407006), containing “UUGCAUGUU” sequence in its loop, and Fox-1 RBD (BBa_K3407004) were first incubated in 1:1 and 1:2 molar ratios shRNA:Fox in binding buffer. 400ng of RNA were loaded in each lane corresponding to 10 fmol, along with 10 fmol of Fox-1 RBD (1:1 ratio) or 20 fmol (1:2 ratio) in a total of 15 μL reaction. Samples were then loaded in a native polyacrylamide gel to avoid protein denaturation.
-
 Figure 3: Illustration of Fox-1 domains used for the assay and their expected interaction with the target sequence of the shRNA or shRNA*. Mutations in either proteins or RNA are depicted as red dots. Mutated sequences of RNA or proteins are colored in orange, while wt protein domains Fox-1 RBD and wt binding sequence in the loop are colored in green.
Figure 3: Illustration of Fox-1 domains used for the assay and their expected interaction with the target sequence of the shRNA or shRNA*. Mutations in either proteins or RNA are depicted as red dots. Mutated sequences of RNA or proteins are colored in orange, while wt protein domains Fox-1 RBD and wt binding sequence in the loop are colored in green.
To demonstrate binding specificity, the same experiment was performed with Fox-1 RBD* (BBa_K3407005), a mutated version of Fox-1 with a reduced affinity for its binding sequence. In addition, the assays were also carried out with shRNA* (BBa_K3407007), a version of the shRNA with two mutations in the loop, which inhibits binding to Fox-1 RBD [1] (Figure 3).
-
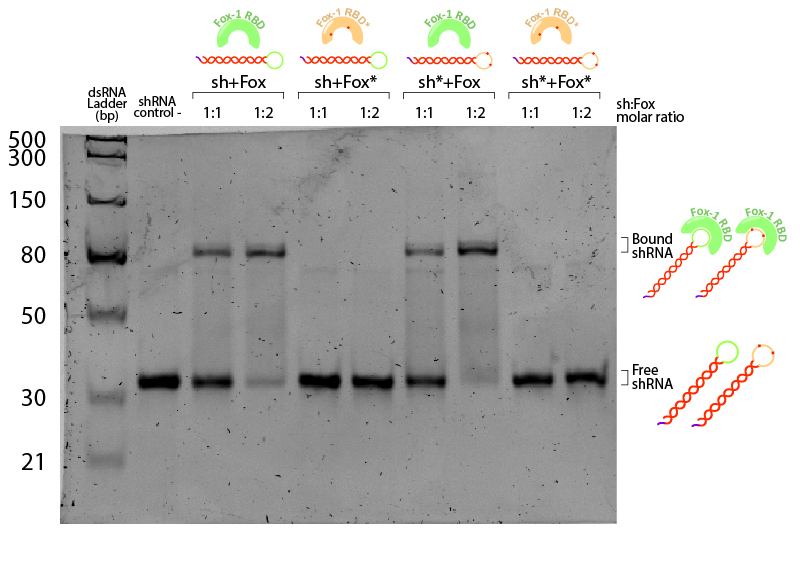 Figure 4: EMSA assay of Fox-1 RBD binding to shRNA. Stars (*) indicate mutated versions of the molecules, where sh* is an shRNA with two mutations in the loop region recognised by Fox-1 RBD (G6A and C3U), and Fox* is the RBD of Fox-1 with two mutations (F160A and F126A) rendering it unable to recognise the RNA target sequence. Control (-) is shRNA incubated in the same conditions without Fox. Incubation was performed in the binding buffer (10 mM Tris pH 7,5; 1 mM EDTA; 100 mM KCl; 0.1 mM DTT; 5% Glycerol; 0.01 mg/ml BSA) in different molar ratios at room temperature for one hour. Electrophoresis with 19:1 (Acrylamide:Bisacrylamide) 12% gel in TAE buffer. Electrophoresis was run at 120 V for about 3 hours in a TAE 1x buffer, stained with Sybr Safe staining solution (0.1% vol./vol. Sybr Safe in TAE buffer).
Figure 4: EMSA assay of Fox-1 RBD binding to shRNA. Stars (*) indicate mutated versions of the molecules, where sh* is an shRNA with two mutations in the loop region recognised by Fox-1 RBD (G6A and C3U), and Fox* is the RBD of Fox-1 with two mutations (F160A and F126A) rendering it unable to recognise the RNA target sequence. Control (-) is shRNA incubated in the same conditions without Fox. Incubation was performed in the binding buffer (10 mM Tris pH 7,5; 1 mM EDTA; 100 mM KCl; 0.1 mM DTT; 5% Glycerol; 0.01 mg/ml BSA) in different molar ratios at room temperature for one hour. Electrophoresis with 19:1 (Acrylamide:Bisacrylamide) 12% gel in TAE buffer. Electrophoresis was run at 120 V for about 3 hours in a TAE 1x buffer, stained with Sybr Safe staining solution (0.1% vol./vol. Sybr Safe in TAE buffer).
The non-bound shRNA and shRNA* appear on the gel as ~30bp dsRNA (negative control, Figure 4). Incubation of shRNA and ahRNA* with wild-type Fox-1 RBD leads to a notable shift of the RNA band at both 1:1 and 1:2 molar ratios. This upper band corresponds to the shRNA-Fox-1 RBD complex, showing that both purified compounds are active. The lower fraction of free shRNA in the 1:2 molar ratio indicates that increasing the concentration of Fox-1 RBD yields a higher amount of complex formed.
-
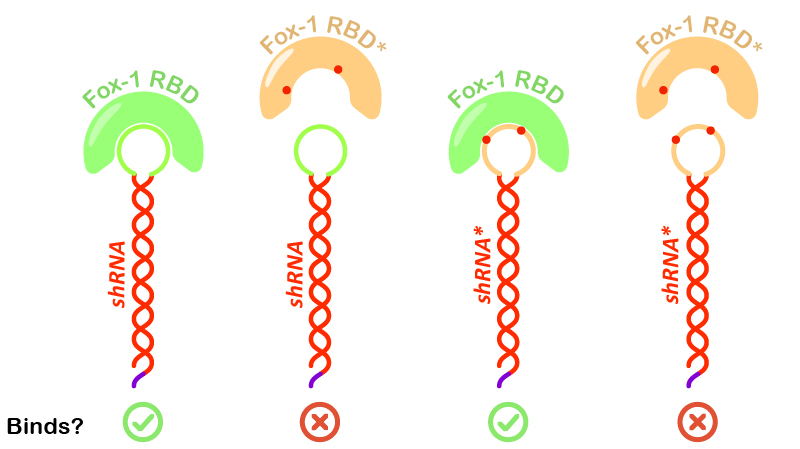 Figure 5: Illustration of Fox-1 domains used for the assay and their obtained results from interaction assay with the target sequence of the shRNA or shRNA*. Mutations in either proteins or RNA are depicted as red dots. Mutated sequences of RNA or proteins are colored in orange, while wt protein domains Fox-1 RBD and wt binding sequence in the loop are colored in green.
Figure 5: Illustration of Fox-1 domains used for the assay and their obtained results from interaction assay with the target sequence of the shRNA or shRNA*. Mutations in either proteins or RNA are depicted as red dots. Mutated sequences of RNA or proteins are colored in orange, while wt protein domains Fox-1 RBD and wt binding sequence in the loop are colored in green.
Interestingly, incubation of shRNA with mutated Fox* RBD does not lead to complex formation. This results corroborates literature data showing that the introduced mutations strongly reduce the affinity of the protein to the target RNA sequence. Surprisingly, Fox-1 RBD (denoted as Fox), but not Fox-1 RBD* (denoted as Fox*), is also able to bind to mutated shRNA*, suggesting that under the studied conditions the mutations in the RNA loop sequence do not abolish complex formation (Figure 5). Different results were previously obtained using surface plasmon resonance [1], which may be explained by differences in the experimental conditions.
References
| None |
