Part:BBa_K3332051
J23100-RBS-GOX-his-tag-terminator
The enzyme catalyzes the reaction of degradating glyphosate to form glyoxalic acid and AMPA and add his-tag to purify the protein. We use K880005 to construct the expression system and to express and to purify the protein.
Biology
Ice nucleoprotein is an anchor protein from Pseudomonas syringae. It can anchor its passenger protein to the cell membrane. N and C terminal of Ice nucleoprotein, which is named after INPNC, can also anchor passenger protein fused with it to the cell membrane. GRHPR, a glyoxylate reductase from human liver, can reduce glyoxylic acid when NADPH is used as cofactor. GRHPR is fused at N terminal with INPNC so that GRHPR can be displayed on the surface of E. coli.[1][2]
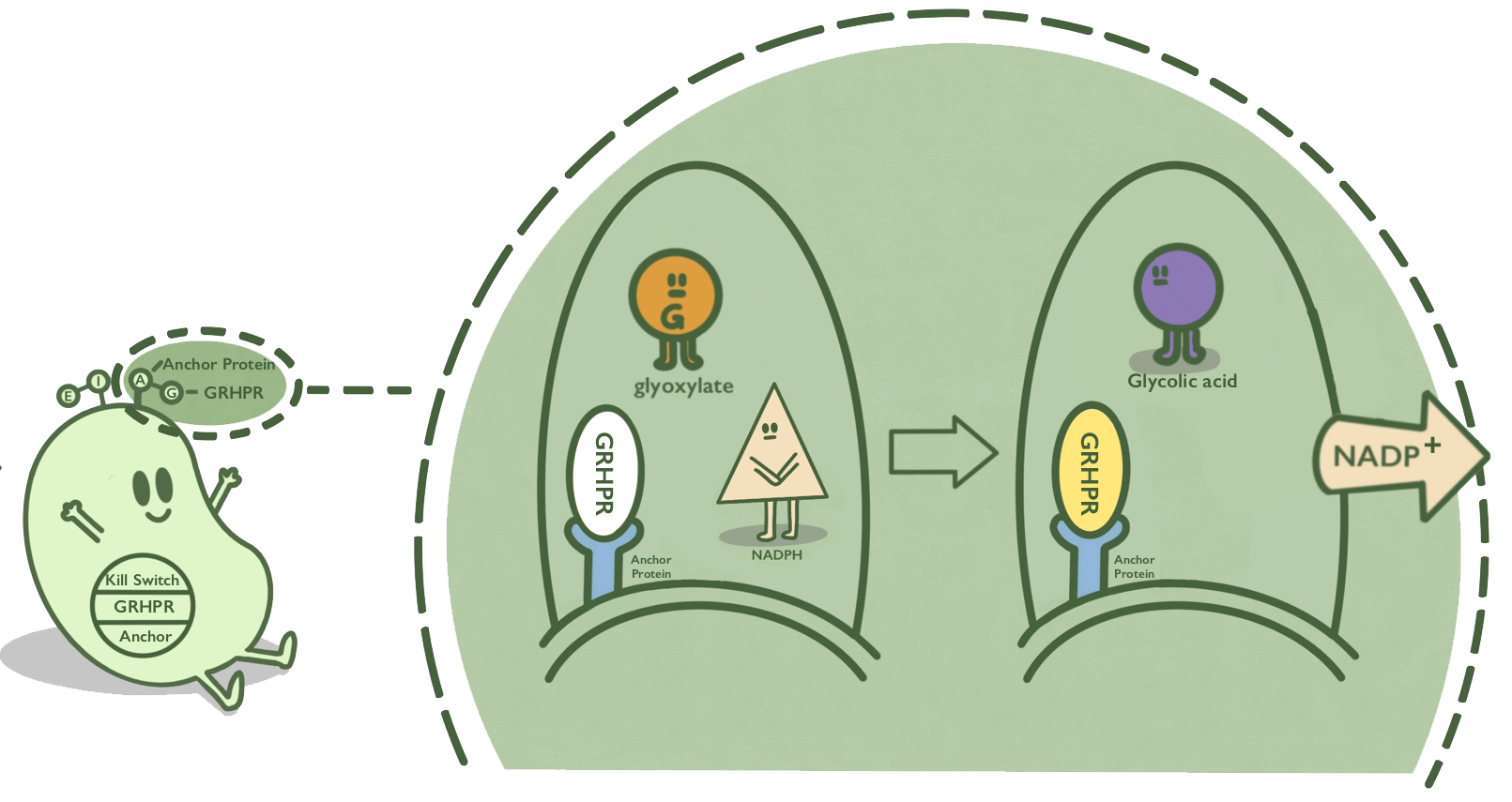
- Fig 1. Mechanism of GRHPR on the surface of E. coli.
Usage
Here, we used BBa_K880005 to construct the expression system and demonstrated the effect of INPNC-GRHPR on the surface of E. coli. We obtained the composite part BBa_K3332057 and transformed the constructed plasmid into E. coli BL21 (DE3) to verify its expression. The positive clones were cultivated.

- Fig 2. Gene circuit of INPNC-GRHPR.
Characterization
1. Identification
After receiving the synthesized DNA, PCR was done to certify that the plasmid was correct, and the experimental results were shown in figure 3.

- Fig 3. DNA gel electrophoresis of PCR products of INPNC-GRHPR-pSB1C3 (Xbal I & Pst I sites)
2. Ability of consuming NADPH
We mixed glyoxylic acid solution, NADPH solution and bacteria solution carrying INPNC-GRHPR. Then, we immediately measured OD340 changes. TECAN® Infinite M200 Pro was used to detect OD340. And when NADPH is consumed, OD340 declines.
We successfully got OD340-Time curves of GRHPR fused with 4 types of anchor protein. When using INPNC-GRHPR, we could see OD340 decreased as the reaction went on. And by using J23100-RBS (BBa_K880005) and GRHPR-Histag as control, we could find that the slopes of experimental and control curves have a significant difference, which means that our fusion protein INPNC-GRHPR is displayed on the surface and works very well.
The result is shown in figure 4.

- Fig 4. OD340-Time curves of GRHPR fused with 4 types of anchor protein.
References
- ↑ Rumsby G, Cregeen D P. Identification and expression of a cDNA for human hydroxypyruvate/glyoxylate reductase[J]. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression, 1999, 1446(3): 383-388.
- ↑ http://2016.igem.org/Team:TJUSLS_China
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 355
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 574
Illegal AgeI site found at 644
Illegal AgeI site found at 721 - 1000COMPATIBLE WITH RFC[1000]
| None |
