Part:BBa_K3189015
Tet Controlled amilCP
Tet Controlled amilCP
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1758
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 146
Illegal XhoI site found at 139 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 80
A sensitive expression assay was then conducted on the functioning BBa_K3189015 by, growing multiple E. coli BBa_K3189015 transformants in a 96-well plate in varying concentrations of tetracycline labeled in Figure 1. A dark pigment was seen at 100 ng/mL tetracycline, with less colour visible at 50 ng/mL for some of the samples, and no colour visible in the absence of tetracycline. This shows that we have improved BBa_1343002 by introducing BBa_K3189001 upstream to act as its new promoter sequence.
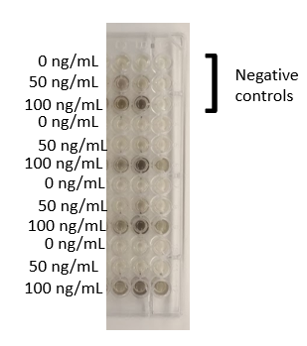
Figure 1: E. coli BL21(DE3) BBa_K3189015 transformants from 11 different colonies grown in LB+tetracycline. The three wells in the top right hand corner are negative controls (E. coli without BBa_K3189015). A dark colour of variable intensity is visible at 100 ng/mL tetracycline for all of the samples.
The construct BBa_K3189015 containing the chromoprotein amilCP (BBa_K1343022) under the control of BBa_K3189001. When the system is induced with 100 ng/mL of tetracycline, a dark blue colour is produced (Figure 2 and Figure 3). This shows BBa_K3189001 is able to function with different reporter proteins other than just gfp.

Figure 2: BBa_K3189015 containing cells in LB broth. amilCP expression being induced using 100 ng/mL tetracycline (left) and uninduced (right).

Figure 3: Pellets of cells of BBa_K3189015 induced and uninduced with tetracycline. Pellet of cells induced with 100 ng/mL tetracycline (left) and pellet of cells uninduced (right).
| None |
