Part:BBa_K3071022
Recombinant Two-component regulatory system RpfG/RpfC construct with sequencing part
Description
This composite part contains the two translational units that encode the two proteins that participate in the two-component regulatory systems RpfC (BBa_K3071018) and RpfG (BBa_K3071019) in the Xanthomononas campestris pv. campestris (Xcc) for sensing diffusible signal factor (DSF). The information encoding for the two related protein is transcribed as one mRNA strand in this design. This part is one of the three parts used to construct our detection system in our part collection, the other two parts include (BBa_K3071023) and (BBa_K3071024).
Biology
RpfC (BBa_K3071000) is a kinase located in the cell membrane of bacteria acting as a sensor. It consists of three domains, including the input domain, histidine kinase (HisKA)domain, CheY-like two-component receiver domain (REC) and the C-terminal histidine phosphotransfer (HPT) domain. The sensory domain is composed of 5 transmembrane helices with periplasmic and cytoplasmic loops less than 20 amino acids long.
RpfG (BBa_K3071002) consists of 2 domains, the REC domain, and the HD-GYP domain, which is a phosphodiesterase involved in the degradation of the second messenger cyclic -di-GMP. When it is phosphorylated by the RpfC, it is activated as phosphodiesterase and alter cyclic-di-GMP level. As a result, it promotes the synthesis of virulence factors, e.g. extracellular enzymes, dispersal of biofilm and motility of the bacterial cell.
Usage

The responsible sensor kinase system detecting DSF in the environment compose of RpfC-RpfG phosphorylation relay. After a series of auto-phosphorylation in RpfC, phosphotransfer from RpfC to RpfG occurs. This leads to the activation of the phosphodiesterase domain in RpfG, which will degrade the second messenger cyclic-di-GMP. As one of the downstream results, Clp binds to its target CBS I and CBS II. Without the DSF induction, cyclic di-GMP will not be degraded but bind to the Clp. The binding between Clp and c-di-GMP prevents Clp from binding CBS.
Using the RpfC-RpfG system as a foundation, we fused the PspF protein with Clp to enhance binding specificity and prevent expression leakage. The DNA binding domain in the C-terminus of pspF (BBa_K3071006) is replaced by Clp to construct a fusion transcription activator for our reporter construct (BBa_K3071024). This transcription activator can activate the CBSI & II-regulated pspA promoter (BBa_K3071014) upon diffusable signal factor (DSF) appearance.
pspA promoter is a sigma-54 (σ-54) regulated activator depending on promoter. sigm54-RNA holoenzyme (σ-54 RNAP) forms an inactive transcriptional initiation complex on this promoter, which can be activated in E. coli by the bacterial enhancer-binding protein PspF (BBa_K3071006). PspF transcription activation domain (pspF TAD) (BBa_K3071008) functions by binding to upstream activation sequences (UAS) near the promoter and contacting the promoter-bound σ-54 RNAP via DNA looping stabilized by integration host factor (IHF). Previous research has demonstrated the property of pspF-dependent and enhancer-specific transcription activation of pspA promoter.
Activation of PspA promoter leads to mRNA expression of the reporter eforRed chromoprotein as an output signal after detecting the existence of DSF.
Characterization


Western Blot data shows that this construct is appliable to correctly express our two candidate proteins under IPTG-induced T7 promoter activity. Our genes sequence is also codon-optimized to be expressed inside Escherichia coli, our RpfG protein (BBa_K3071002) shows a higher rate of expression compared to the originally reported sequence (BBa_K3071003) in western blot result.
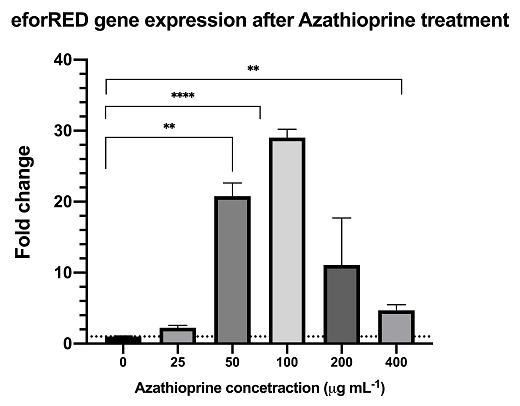
Azathioprine is an indirect supressor to cyclic-di-GMP, treatment of azathioprine to bacteria culture can reduce the cyclic-di-GMP level and lead to activation of the pspF-Clp and by-pass the RpfC/RpfG two-component system.The result from rt-qPCR data shows that the above synthetic biological system is functional with significant up-regulation of reporter mRNA.

The result from rt-qPCR data shows that the above synthetic biological system is functional with significant up-regulation of reporter mRNA upon DSF activation.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 349
Illegal NgoMIV site found at 2647
Illegal AgeI site found at 1181
Illegal AgeI site found at 1660
Illegal AgeI site found at 1744 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 398
Illegal BsaI.rc site found at 262
| None |
