Part:BBa_K2916050
Toehold for Grapevine Endogenous Control 1.1
Usage and Biology
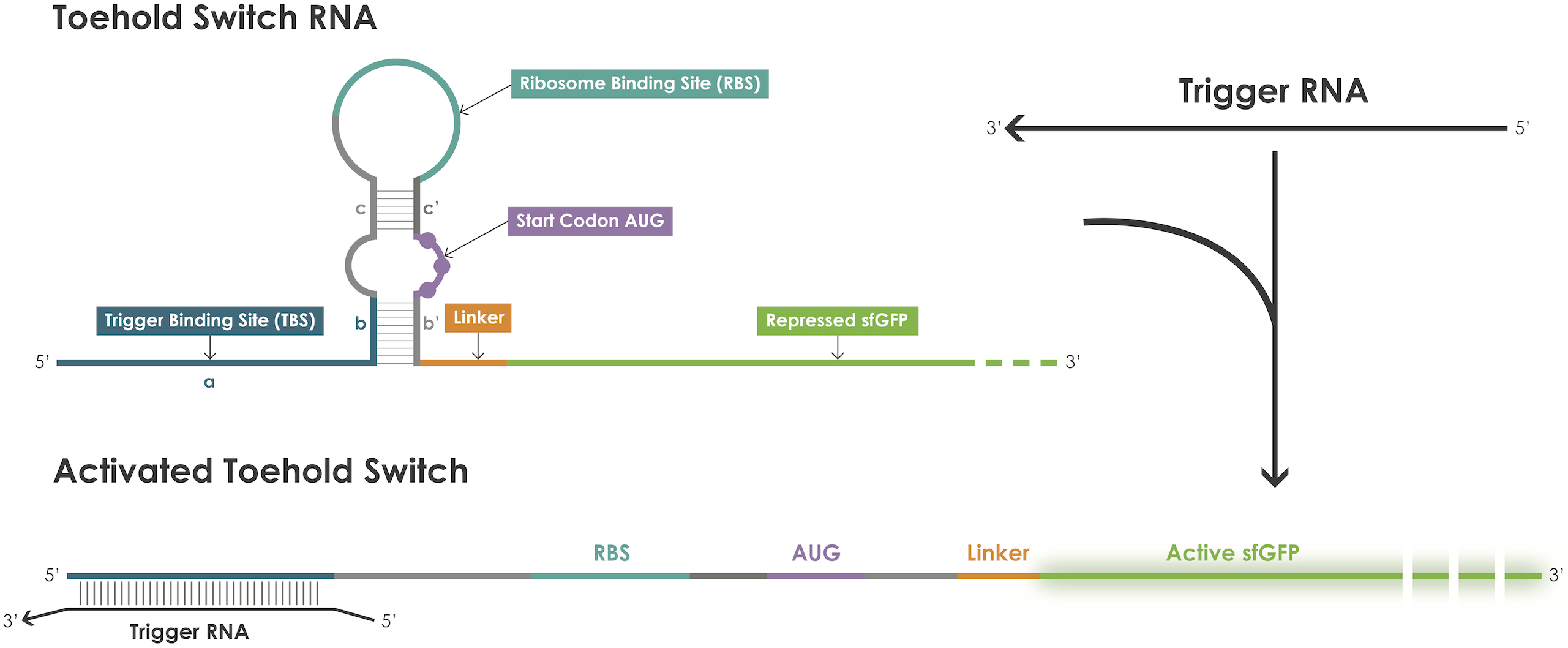
Toehold switches are synthetic riboregulators that regulate the expression of a downstream gene. They sequester the Ribosome Binding Site (RBS) and Start Codon by forming a secondary structure called a hairpin structure in its OFF state (acting as a “lock” for protein expression). A second strand of mRNA, complementary to the trigger binding region, needs to be present to activate the toehold. It will bind to that region and linearize the hairpin structure, freeing the RBS for the ribosomes to bind and allowing for translation of the downstream gene. Our toehold switches function at 37°C, and protein production is carried out in a cell-free PURE system. Reference Toehold switches: de-novo-designed regulators of gene expression, Green et al., 2014 In the encoding part, we have a toehold region, a hairpin region which contains a stem and a loop, a refolding region and a spacer. In design, it is crucial to exclude any ending codon in the refolding and spacer region. The refolding region allows ribosomes to access the protein coding region with less energy, thus increasing the kinetics of the reaction.
Improvement by Evry_Paris-Saclay 2020
This part is a toehold switch sensor [1] (Figure 1) for sequence-based detection of grapevine. It targets a fragment of the TrnL-UAA gene of Vitis vinifera.
Figure 1. Toehold switches principle.
Its secondary structure predicted by NUPACK web server [1] using default parameters is represented in Figure 2 and here-after in dot-bracket notation.
........................(((((((((((((..((((((....(.....)....))))))..)))))))))))))...((((((....)))))).....
Figure 2. Secondary-structure prediction of this part with the ATG of the reporter gene. The prediction was realised using the NUPACK web server [2] with default parameters and graphically represented using the forna RNA secondary structure visualization tool [3]. Nucleotides were coloured to match the different segments in Figure 1.
However, the trigger binding site of this toehold switch is in the same orientation as in the transcriptional orientation of its target gene. Thus it is unlikely that this toehold switch will be able to detect the presence of an RNA naturally expressed in a cell.
This part has been improved to be functional. We have successfully built two parts, the Grapevine Switch 3.1 (BBa_K3453079) and the Grapevine Switch 3.2 (BBa_K3453080), which are able to act as toehold switches and (i) efficiently repress the downstream reporter gene expression in the absence of the cognate trigger and (ii) release the translation inhibition in the presence of the cognate trigger, an RNA molecule having the transcription orientation of the TrnL-UAA gene of grapevine.
References
[1] Green AA, Silver PA, Collins JJ, Yin P. Toehold switches: de-novo-designed regulators of gene expression. Cell (2014) 159, 925-939.
[2] Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, Dirks RM, Pierce NA. NUPACK: Analysis and design of nucleic acid systems. Journal of Computational Chemistry (2011) 32, 170–173.
[3] Kerpedjiev P, Hammer S, Hofacker IL. Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics (Oxford, England) (2015) 31, 3377–3379.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 53
| None |