Part:BBa_K2601021
Tet07-Frb-yEGFP
Introduction
A widespread use of rapamycin, an anti-fungal antibiotic, has been developed to take advantage of the small molecule’s ability to heterodimerize proteins. Rapamycin binds to the FK506 binding protein (FKBP) as well as to a 100-amino acid domain of the mammalian target of rapamycin (mTOR), known as the FKBP-rapamycin binding domain (Frb). Proteins of interest can be expressed as fusions to FKBP or Frb, and then conditionally dimerized by adding rapamycin.
Design
Some membrane-less organelles, such as stress granules and P bodies, have been discovered in recent years. Proteins condense into droplets and assemble these organelles through a process called phase separation. Physically, phase separation is the transformation of a one-phase thermodynamic system to a multi phase system, much like how oil and water demix from each other. According to thermodynamics, molecules will diffuse down the gradient of chemical potential instead of concentration. This is exactly why proteins will self organize into granules, diffusing from regions of low concentration to regions of high concentration. Here is an illustration of phase separation in cells.
 |  |  |
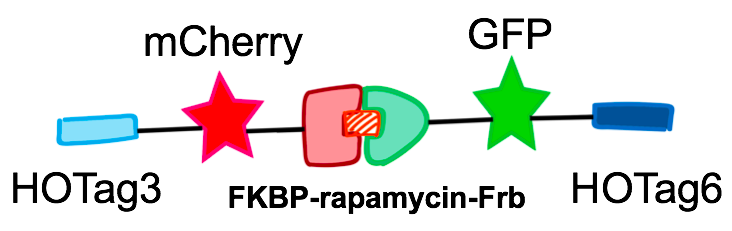
In order to rationally design a synthetic organelle based on protein phase separation, we needed a multivalent module and a protein-protein interaction module. The paired FKBP and Frb was one of the bioparts that we chose to introduce protein-protein interaction. FKBP and Frb could dimerize upon adding rapamycin. As for the multivalent module, we turned to de novo-designed homo-oligomeric short peptides. These short peptides are called HOTags (homo-oligomeric tags). HOTags contain approximately 30 amino acids. They have high stoichiometry, forming hexamer or tetrameric spontaneously.
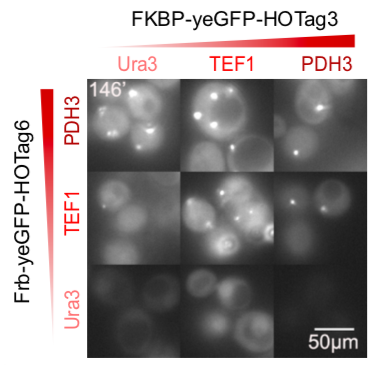
The hexameric HOTag3, together with the tetrameric HOTag6, could robustly drive protein phase separation upon protein interaction (achieved by FKBP-Frb module). To verify the feasibility of the system, we fused two fluorescence proteins with the two components of synthetic organelles. Thus, we could observe the self-organization of components and the formation of organelles under fluorescence microscope. We named our system SPOT (Synthetic Phase separation-based Organelle Platform) because it could form granules (fluorescent spots) in yeast. Here is a demonstration of our overall design.
Properties
The interaction between FKBP and rapamycin has been well characterized (Kd ≈ 0.2 nM), and early experiments suggest that formation of a ternary complex including FRB is quite favorable (Kd ≈ 2.5 nM). FKBP and Frb do not interact in the absence of rapamycin.
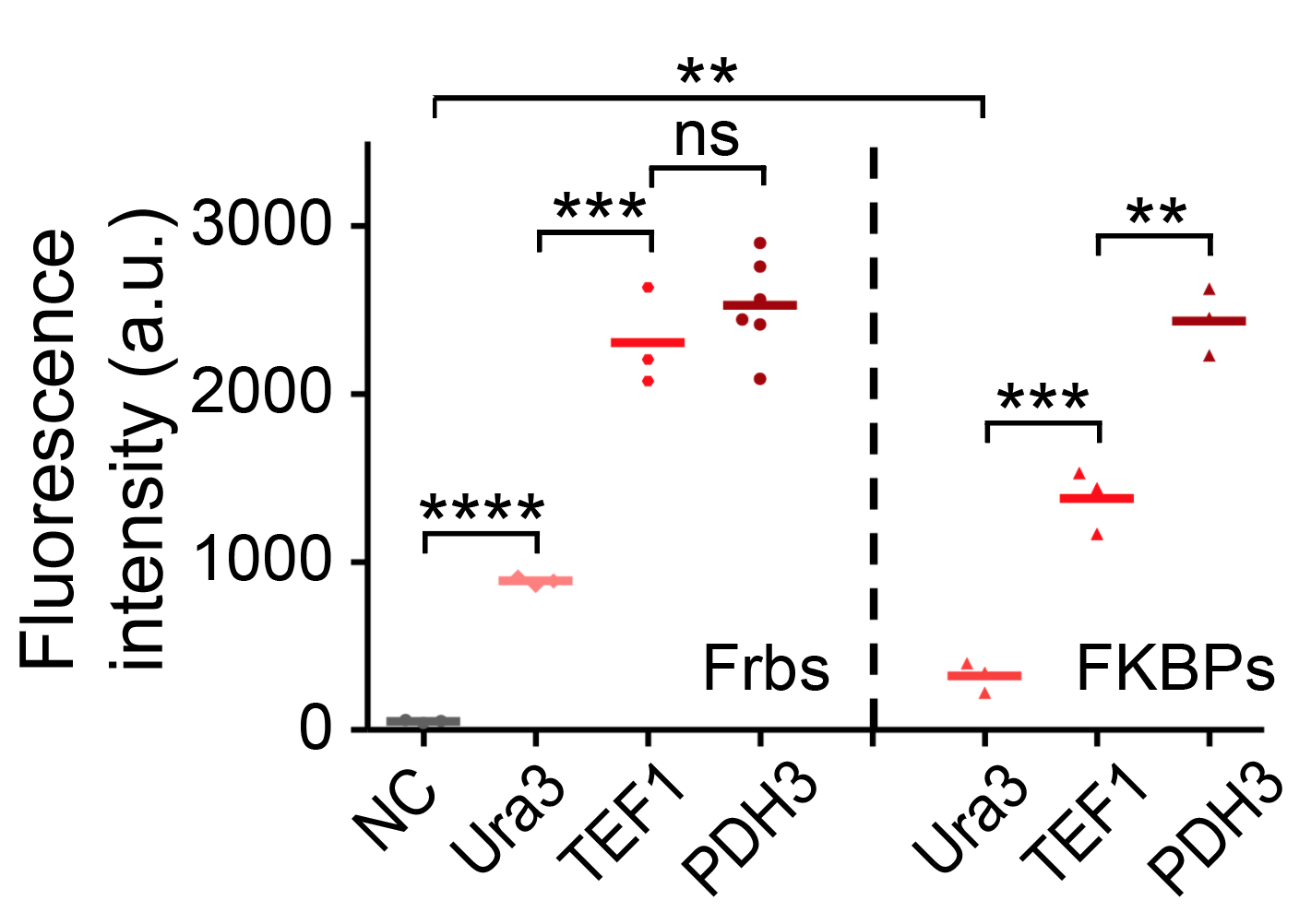
Tet07 is a inducible yeast promoter, which can be induced by doxycycline. We have tested the strength of Tet07 using flow cytometry.
Frb-yeGFP has successfully expressed in yeasts.
Our results confirmed that FKBP/Frb module fused with HOTags can drive phase separation in yeast.
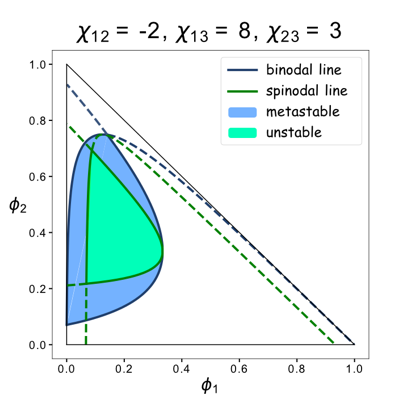
We set up a thermodynamic model characterizing our system. Phase separation is easy to take place in the green area, where the system is unstable, while it can also happen in the blue area, where the system is metastable. Phi 1, phi 2 and phi 3 represent the volume fraction of FKBP, Frb and water, respectively. The parameter chi represents the interaction energy. Chi one three can be roughly interpreted by the interaction strength between FKBP and water, while chi two three indicates the interaction strength between Frb and water. Similarly, chi one two denotes the interaction between FKBP and Frb. Whether chi one three is equal to chi two three decides whether the diagram is symmetric. This is a useful instruction for our experiments. To some extends it can save us from some unnecessary trials. In our experiment, we chose promoters with different strength to adjust volume fraction of FKBP and Frb. Phase separation could be observed only when FKBP-HoTag3 had a low level expression while Frb-HoTag6 had a high level expression. The experimental results was consistent with the model.
| | |
Here are the kinetic simulation results. The larger chi is, the sooner the system relaxes into equilibrium state, which means the faster the phase separation appears. In our experiment, we changed the interaction energy between FKBP and Frb by changing the concentration of rapamycin. Higher concentration of rapamycin led to faster SPOT formation, which was also consistent with the model.
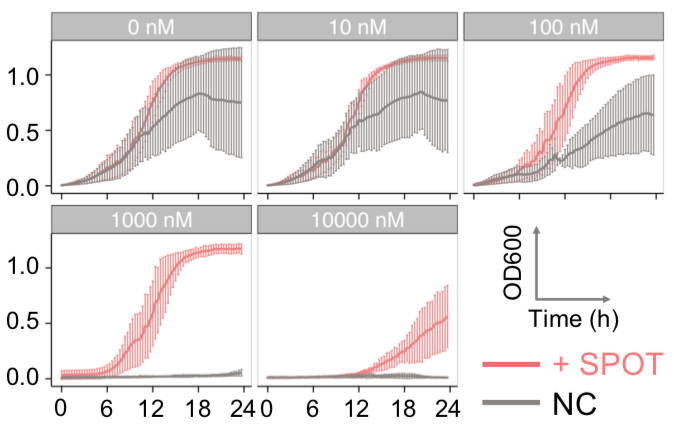
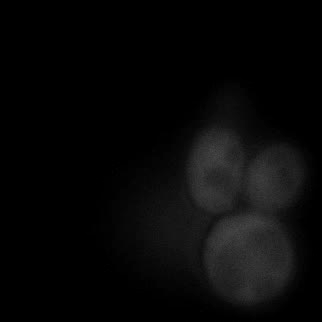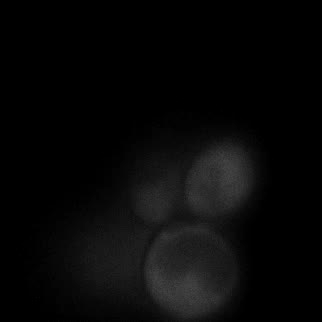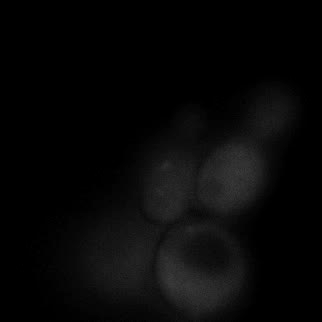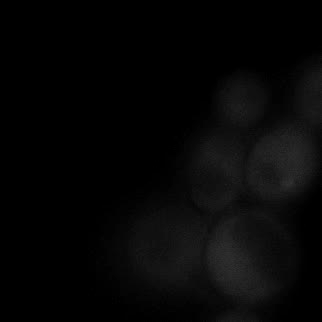 | 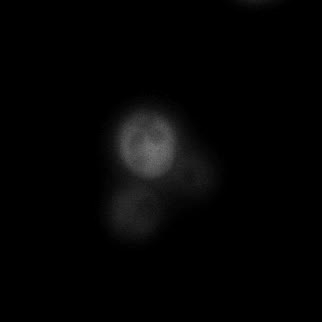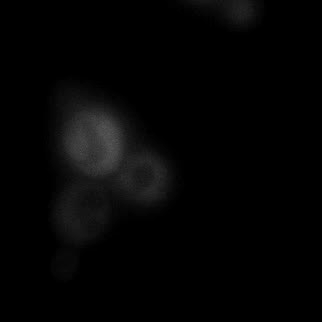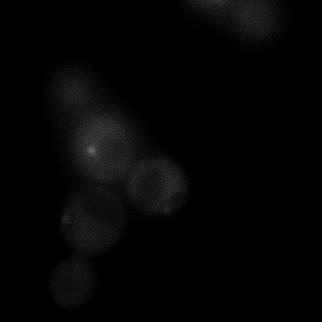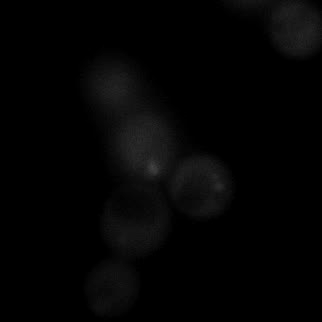 | 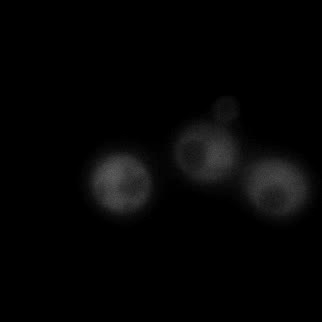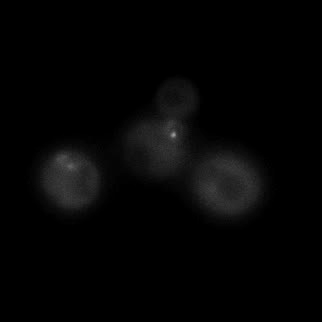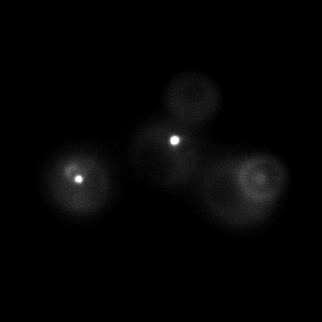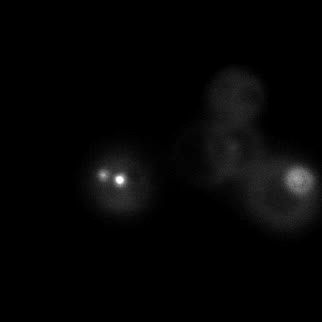 |  |
| Figure 11. Yeasts with FKBP-HOTag3 and Frb-HOTag6 were induced by different concentrations of rapamycin. | |||
FKBP-Frb based phase separation system could alleviate the inhibitory effect of rapamycin. It could sequester rapamycin in the granule and rescue the yeast from the toxicity of the rapamycin. From the yeast growth curve we could see that there was a significant difference between cells with and without phase separation.
Improve BBa_K209496
Improve https://parts.igem.org/Part:BBa_K209496
We characterized the expression of Frb-yEGFP with promoter Tet07 and measured expression through flow cytometry. Promoter Tet07 can be induced by Dox.
Please view http://2018.igem.org/Team:Peking/Improve for more details.
Reference
1. Putyrski, M., & Schultz, C. (2012). Protein translocation as a tool: The current rapamycin story. FEBS letters, 586(15), 2097-2105.
2. Banaszynski, L. A., Liu, C. W., & Wandless, T. J. (2005). Characterization of the FKBP Rapamycin FRB Ternary Complex. Journal of the American Chemical Society, 127(13), 4715-4721.
3. Zhang, Q., Huang, H., Zhang, L., Wu, R., Chung, C. I., Zhang, S. Q., ... & Shu, X. (2018). Visualizing Dynamics of Cell Signaling In Vivo with a Phase Separation-Based Kinase Reporter. Molecular cell, 69(2), 334-346.
4. Woolfson, D. N., Bartlett, G. J., Burton, A. J., Heal, J. W., Niitsu, A., Thomson, A. R., & Wood, C. W. (2015). De novo protein design: how do we expand into the universe of possible protein structures?. Current opinion in structural biology, 33, 16-26.
5. Berry, J., Brangwynne, C. P., & Haataja, M. (2018). Physical principles of intracellular organization via active and passive phase transitions. Reports on Progress in Physics, 81(4), 046601.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1010
Illegal BamHI site found at 1031 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1677
Illegal SapI.rc site found at 919
//chassis/eukaryote/yeast
//proteindomain/binding
| biology | Saccharomyces cerevisiae |