Part:BBa_K2546001
2-haloacid dehalogenase(HAD)
This part is coding for 2-haloacid dehalogenase, which can cleavage carbon-halogen bonds of halogenated compounds.
RESULT
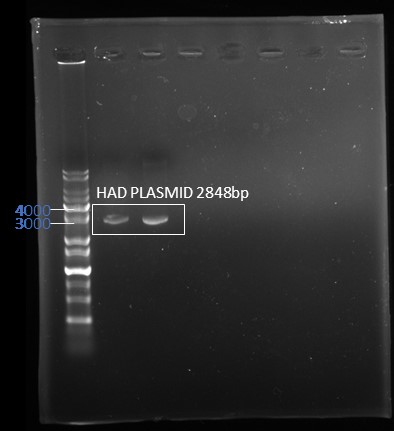
Through the 3A assembly, we successful assemble the HAD in the pSB1C3 vector. First, we amplify HAD through the PCR and through the gel electrophoresis to check the correct size.
Then clean up the PCR product and cut the HAD with EcoRI and PstI, and cut the pSB1C3 with EcoRI and PstI. Then another clean up for restriction enzyme product. Add ligase and buffer after calculate the nucleic acids concentration of the HAD and pSB1C3 and the ratio of the insert : vector. And the ligation is under 4℃ overnight. After the transformation, you can collect the colony overnight, and incubate in the LB to check if it is correct in the next day.
This is the result through the plasmid extraction, and the target place is at 2848bp, which is match to the result.
And also check again through the restriction enzyme digestion. The picture below is be cut by EcoRI and PstI. You can see there are two band, one is in 2070bp which refer to pSB1C3 and the other one is in 819bp, which is HAD.
Contributions
Group: iGEM22_WVHS_SanDiego
Authors: Ananya Bharathwaj, Madison Yang
Summary: Haloacid dehalogenase (HAD) is an enzyme that has been shown to cleave carbon-halogen bonds. For our project, we modeled the interaction between PFOA and HAD (derived from Xanthobacter autotrophicus), and the conformation at the maximum binding energy (binding energy: -5.8, ligand efficiency: 0.23). For in-lab work, we took the sequence of HAD, codon-optimized it for E-coli, and made it compatible with the Gibson and Golden Gate assembly methods of our u loop system (removing BsaI and Sapl restriction sites, adding 20 bases of homology to the each end of the gene sequences, and adding C-D overhangs).
Below are images of our modified sequences developed in SnapGene, along with our protein model developed from Autodock 4.


Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 684
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 333
Illegal NgoMIV site found at 430
Illegal AgeI site found at 673 - 1000COMPATIBLE WITH RFC[1000]
| None |