Part:BBa_K2323004
Lwa Cas13a under T7 promoter with solubility tag
Uses a T7 promoter and RBS B0034 (BBa_B0034) to express Lwa Cas13a (BBa_K2323000) flanked by a 6x His-Tag to allow purification and a SUMO-tag to improve solubility. This is terminated by a Tphi terminator. The whole coding sequence is also available as BBa_K2323001.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1768
Illegal BglII site found at 3448
Illegal BamHI site found at 144 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 2847
Usage and Biology
Cas13a from Leptotrichia wadei (Lwa) is class 2 type VI CRISPR-Cas effector. It is capable of targeting specific ssRNA and cleaving them using its bounded crRNA as guide strand. Unlike other ssRNA CRISPR-Cas effectors, Cas13a is capable of processing the precursor crRNA (pre-crRNA) by itself, cleaving, binding the RNA sequence and then building the Cas13a:crRNA surveillance complex. One characteristic of Cas13a along with other type VI members is its ability to degrade other RNA strands regardless of their sequences, provided the protein has been activated by binding and digesting the target sequence complementary to the crRNA. This "collateral activity" makes Cas13a a powerful biological tool, as unspecific RNA degradation can be triggered on command by providing the target RNA.
Plasmid composition
The Biobrick contains a promoter, RBS B0034 and Lwa-Cas13a with a His-SUMO-Tag, that allows for easy expression and purification. The SUMO-tag increases solubility and can be cleaved off with a SUMO-protease after the affinity purification.
Cloning, Expression and Purification
Cloning
We ordered Lwa in 2 gblocks from IDT, ready for Golden-Gate-Assembly. We ligated it into the pSB1C3-ccdB by restriction digestion of the backbone and having adequate BsaI sites within the gblocks. We confirmed the cloning by colony-PCR, analytical restriction digest and sequencing at GATC. The colony was stored as a Cryo-stock at -80 °C.

| Name | 5'-3' primers sequences |
|---|---|
| pseq-Lwa-01 | CGTTTTCTGTATGACGGCATCC |
| pseq-Lwa-02 | TCCGGATATGAGCGAACTTAAGA |
| pseq-Lwa-03 | GAGAACCTGAAGATGTTTTA |
| pseq-Lwa-04 | ATCATAGGAAAGCAGTTTG |
Expression
As the sequence was ordered codon optimized, we could express it in BL21 star. Expression was done overnight at 16 °C in 2xYT media in the pressence of 1 mM IPTG.
Purification
Characterisation
Target dependent activity of Cas13a Lwa
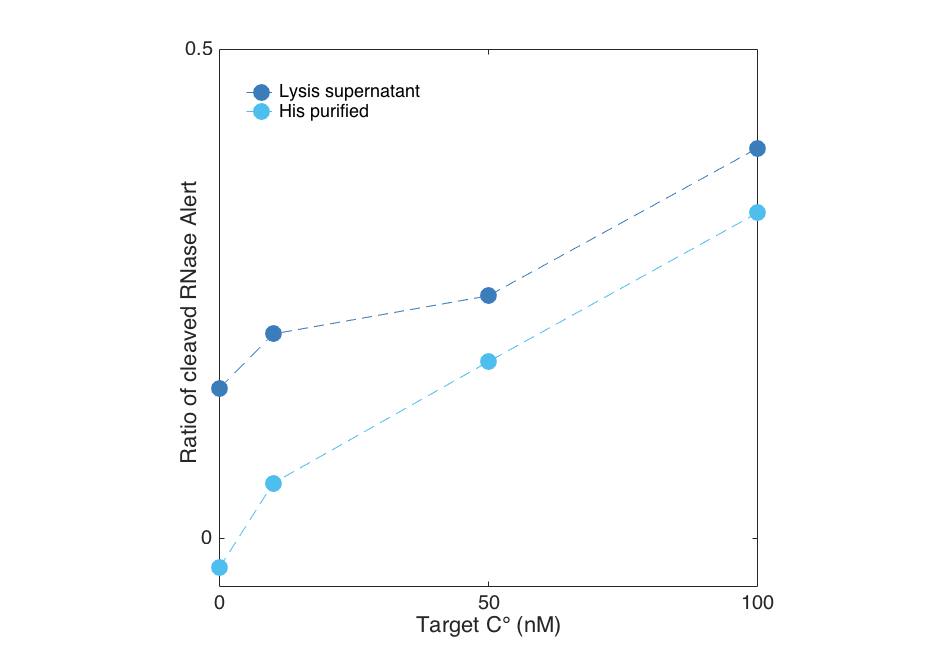
We show below, that Cas13a gets activated upon target binding in a concentration dependent manner. In this case, we detected a fragment of the 16S rRNA of E. coli in a plate reader experiment by using the RNaseAlert system readout. After less than 30 minutes a difference between a target containing sample and the negative control (sample without Cas13a Lwa) is visible for both the purified and the unpurified Cas13a. However, the unpurified Cas13a has a visibly larger activity at low target concentrations, and even shows cleavage at 0 nm target. This is probably attributed to residual RNases that are within the cellular lysis mix. The positive control contained RNAse A, which cleaved all RNAseAlert before the measurment could be started in the plate reader.
- Target dependent activity screen of the purified Cas13a
When plotted against each other, the previously mentioned residual activity becomes even clearer. For this reason we suggest a proper purification of the protein to get reproducible results.
Contribution by Team ZJUT_China_B 2020
Group: Team ZJUT_China_B 2020
Author: Xiaojie Zhou
Summary: We learned from literature that Cas13 protein homologues not only have target dependent activity(characterized by iGEM 2017_Munich) but also have different motif preference upon specific nucleotides and LwaCas13a protein's base preference is poly U/AU.This year,on the basis work of iGEM 2017_Munich, ZJUT_China_B further characterized the base preference of LwaCas13a protein's collateral cleavage,which may be a potential feature to achieving multivirus detecting in one pot.
Methodology
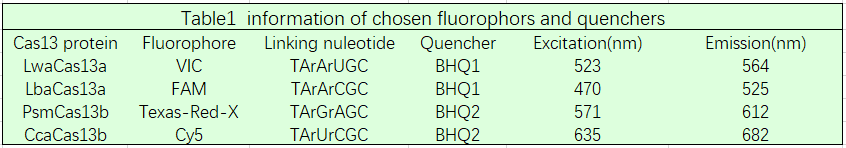
Based on our multivirus detecting goal, we designed four kind fluorescent reporters (Table 1). The linking nucleotide are AC, AU, GA, and UC respectively. The fluorophore are FAM, Cy5, Texas-Red-X and VIC respectively. This enables us detecting the corresponding extention of the cleavage. We added those 4 fluorescent reporters in one detecting pot, and got the following data.
Results
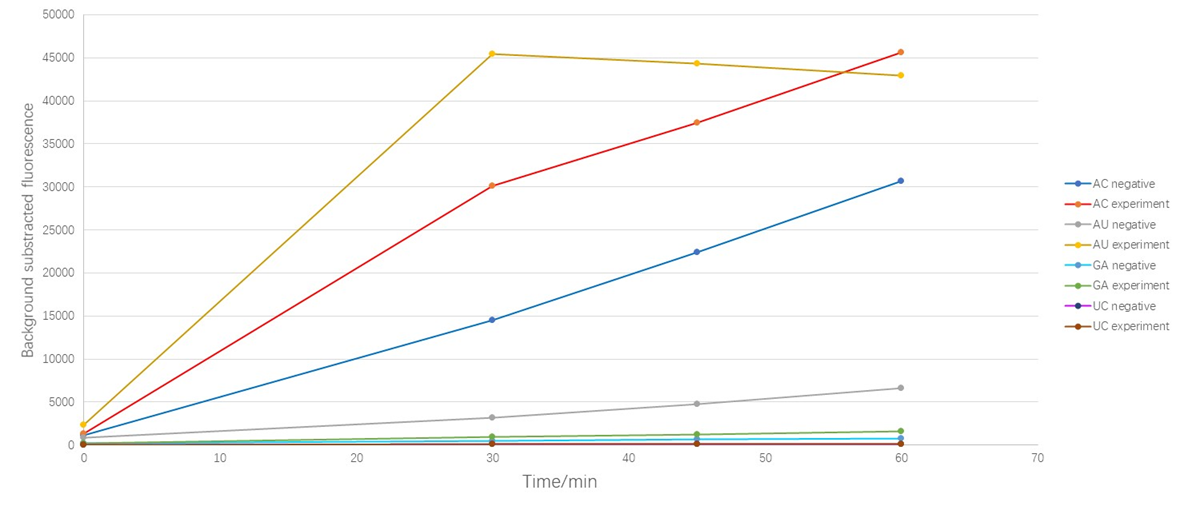
1. Because our former experiment shows that as the excitation goes on, our fluorescent dyes show a certain degree of photobleaching phenomenon, so we decrease the time of excitation. For details of photobleaching phenomenon,click ZJUT_China_B Model
2. The "negative" is the group that we didn't add target while the "experiment" is the group where we add our target.
Analysis
As shown in Figure 1, the results suggest that LwaCas13a protein could cleave AU and AC oligonucleotide even though there is no target in the detecting system which we called "the leakage acticity of Cas13 protein ". Obviously, the LwaCas13a protein shows more "leakage" upon AC while the leakage upon GA and UC are quite slight. When we added target sequence in the detecting system, the cleavage of AC obviously increased (the RFU increased 2.04 times at 30min) but the background subtracted fluorescence of AU reporter increased in a cliff like manner (the RFU increased 13. 89 times at 30min), which means that when the target was added, the protein's activity to cleave AU was activated significantly. Due to the preference appears only when the target was added, we defined this as the cleavage base preference of LwaCas13a protein, or more specifically, "the activating associated cleavage base preference " of the protein.
Contribution by Team IISER_Kolkata 2021
Group: iGEM21_IISER_Kolkata
Author: Soumi Bhattacharyya, Shubhamay Das
Summary: We learnt that Team iGEM17_Munich has performed the assay for target-dependent activity of purified Cas13a protein under different concentrations of target RNA molecules. Team ZJUT_China_B 2020 has characterized the base preference of collateral cleavage activity of LwaCas13a protein. Due to limitations in laboratory access, we have contributed more information about this part through literature survey.
New information: Zhang Lab devised the SHERLOCK technique which uses isothermal preamplification with specific activation and non-specific cleavage activity of Cas13a to detect small RNA or DNA molecules. This is a significant scientific advancement in the field of rapid nucleic acid detection used in diagnostics and biotechnological applications. LwaCas13a collateral cleavage activity results in the production of cleaved RNA products with hydroxylated 5’ ends and 2’-3’ cyclic phosphate ends. A Type III CRISPR Cas system protein called Csm6 gets efficiently activated by 2’-3’ cyclic phosphate terminated hexadenylated ends [1].
Zhang et al. have further devised SHERLOCKv2 [2] which is an advancement of the SHERLOCK technique [3]. In this method, orthogonal CRISPR enzymes Cas13a and Csm6 are multiplexed together to detect RNA molecules. SHERLOCKv2 acts by the method of initial specific activation of cas13a by target RNA molecules which then goes on to generate cleavage residues/overhangs of 2’-3’ cyclic phosphate by collateral cleavage of RNA in the sample. These overhangs further activate Csm6 which also carries out collateral cleavage. According to literature, it was found that this can be used to detect as low as 2 attomolar input RNA with a preamplification step (Eg: Recombinase Polymerase Amplification). They also devised a lateral flow detection technique where the signal of LwaCas13a is enhanced by EiCsm6(Csm6 from Enterococcus italicus) solely, without the preamplification step by RPA.


References
1) Niewoehner, O., Garcia-Doval, C., Rostøl, J. et al. Type III CRISPR–Cas systems produce cyclic oligoadenylate second messengers. Nature 548, 543–548 (2017). https://doi.org/10.1038/nature23467
2) Jonathan S. Gootenberg, Omar O. Abudayyeh, A Max J. Kellner, A Julia Joung, A James J. Collins, A Feng Zhang et al. T Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. 2018 Science P439-444 V 360 N 6387 doi:10.1126/science.aaq0179
3) Kellner, M.J., Koob, J.G., Gootenberg, J.S. et al. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc 14, 2986–3012 (2019). https://doi.org/10.1038/s41596-019-0210-2
4) http://2017.igem.org/Team:Munich
5) https://www.uniprot.org/uniprot/U2PSH1
Contribution by Team GO-Paris-Saclay 2021
Group: Team GO-Paris-Saclay 2021
Author: Enzo Garcia, Florent Poubanne
Summary:
Cas13a is a RNA-dependent RNAse from the Gram-negative bacterium Leptotrichia buccalis belonging to the class VI CRISPR-Cas systems measuring 1159 amino acids. Its activation by hybridization of a crRNA with which it is associated with its complementary single-stranded RNA allows it to cleave any single-stranded RNA in an aspecific manner, this phenomenon is called collateral activity. This collateral activity allows the use of this protein for the detection of RNAs (here miRNAs) using a fluorescent single-stranded RNA reporter.
Methodology
Production and purification
For the production of Cas13a, E.coli Rosetta strain was transformed with plasmids pC0072 LbuCas13a His6-TwinStrep-SUMO-BsaI (Gootenberg et al., 2018). The plasmids, which were ordered from Addgene, carry an ampicillin resistance gene and allow the constitutive production of the transcription inhibitor LacI. Expression of the gene of interest is under the control of the T7 polymerase (already expressed by Rosetta strain) promoter and the lacO operator. Transformed bacteria were grown overnight in LB supplemented with ampicillin (200 mg/mL), chloramphenicol (30 mg/mL) and glucose (0.2%). Cells were diluted in 1 L Terrific Broth (rich medium) supplemented with antibiotics to OD600=0.1 and grown at 37°C until the OD reached 0.5. Then, IPTG (0.5 mM) was added and the culture was shaken at 18°C for 16 h for protein production. The bacteria were harvested by centrifugation at 5000xg for 15 min and resuspended in 40 mL LEW buffer (50mM NaH2PO4 300 mM NaCl pH 8.0) containing Complete protease inhibitor cocktail (Roche). Following lysis by sonication, the proteins carrying His-tags were purified using nickel columns (Protino Ni-IDA 1000, Machery-Nagel) and digested with SenP2 (Cas13a) or in order to remove the histidine tags and solubilization domains they carried. Cas13a was then purified by FPLC on heparin column (1 mL, GE Healthcare) using a gradient using solution A (50 mM NaH2PO4 pH 7.5, 100 mM NaCl, 1 mM beta-mercaptoethanol (BME)) and solution B (50 mM NaH2PO4 pH 7.5, 1 M NaCl, 10% glycerol, 1 mM BME). Proteins were then diluted 3-fold in 50 mM NaH2PO4 pH 7.5, 20 m M NaCl, 10% glycerol, 1 mM BME. Finally, a Millipore centrifugal device was used to concentrate the protein. Proteins were stored at -20°C after adding glycerol up to 50%. Proteins were analyzed by SDS-PAGE and Coomassie staining.
miRNA detection assays
Cas13a protein coupled with its crRNA had been incubated with bta-miR-7863 or miR-125b miRNAs and a ssRNA reporter. The miRNA hybridized with the crRNA and then activated the Cas13a’RNAse activity allowing it to cleave the reporter which, then, emitted fluorescent signal. Thus, it was possible to follow the detection of the miRNA in a quantitative way by following the fluorescence level.
Results
Production and purification
Non-digested Cas13a measuring 155 Kda and digested Cas13a measuring 138 Kda we can conclude that Cas13a has been efficiently digested and its tags and solubilization domains removed.
Following the purification on heparin column we could observe 1 absorbance peaks at 280 nm at 50 minutes corresponding to Cas13a with tags removed.
miRNA detection assays
Referring to the results presented in Figures 3 and 4, the optimal conditions for miR-125 detection with Cas13a appear to be a crRNA concentration of 5nM and a 60-minute pre-incubation (Fig 3C and 4). Indeed, although the highest fluorescent signal is observed with a 30-minutes pre-incubation (Fig 3B), the corresponding profile at the other concentrations suggests that this is a result due to measurement or handling error. On the other hand, we can see that a pre-incubation of 10 minutes or 120 minutes seems to result in a low level of fluorescence, even for the most optimal crRNA concentrations (Fig 3A and D).
The results presented in Figures 5 and 6 seem to correlate with those presented in Figures 3 and 4. As with miR-125 detection, the conditions for obtaining an optimal signal when detecting miR-7863 by Cas13a appear to be 5nM of crRNA and 60 minutes of pre-incubation (Fig 5C and 6).
The results presented in Figure 7 suggest that the optimal concentration of miRNA for detection of miR-125 by Cas13a is 50 nM. The limit of detection appears to be 20nM.
By observing the results in figure 8 we can conclude that the activation of Cas13a is well specific. Indeed, Cas13 coupled to crRNA-125 does not seem to cleave the fluorophore when incubated with non-specific RNA (bacterial or human) or miR3613.
Conclusions
In conclusion, we succeeded in producing and purifying Cas13a. The tests shows that while the specificity of the Cas13a1 reaction is high as we were able to detect miRNA 125 (Fig 3-5 and 7) and 7863 (Fig 5-6) without detecting non-specific RNAs (Fig 8), the sensibility needs to be improved and incubation times need to be tested over a wider range of durations to further improve the cleaving response.
| None |