Part:BBa_K2185003
Penicillium alpha amylase w/ no promoter
This composite part, designed and submitted by the 2016 Broad Run iGEM Team, contains an gene from the fungus Penicilium that codes for an alpha amylase enzyme. This composite part contains the coding sequence, kozak sequence, a mating factor secretion tag, and a terminator. It does not include a promoter. The amylase enzyme can be expressed by cloning this part into a plasmid with a promoter built in (such as the S.cerevisiae yeast vector pAG36).
Components
Kozak Sequence - BBa_K165002
This kozak sequence functions as the ribosome binding site for S.cerevisiae. It is involved in translating the sequence from mRNA to amino acids.
Mating Factor Secretion Tag - BBa_K792002
This part is the secretion signal peptide from the S.cerevisiae α-mating factor. This signal peptide directs the secretion of the produced protein, allowing the protein to interact with the environment outside the cell.
Fungal Alpha Amylase Coding Sequence - BBa_K2185002
This coding sequence codes for the alpha amylase enzyme from the Pencillium fungus. The enzyme can break alpha bonds linking polysaccharides, thus degrading starch into monosaccharides.
ADH1 Terminator - BBa_K392003
This is the terminator region from yeast alcohol dehydrogenase (ADH1) gene.
Proof of Expression and Function=
For testing this part, we cloned this composite part into a S.cerevisiae vector that contained a TEF1 constitative promoter and transformed this vector with the insert in to S.cerevisiae. We combined liquid cultures of our genetically modified S.cerevisae with a starch solution, to determine if the cells were producing the amylase enzyme, and if the enzyme was functioning properly and degrading starch. We found that the cells were indeed producing a functional amylase, evidenced by the significant amount of starch degraded in a period of 6 hours.
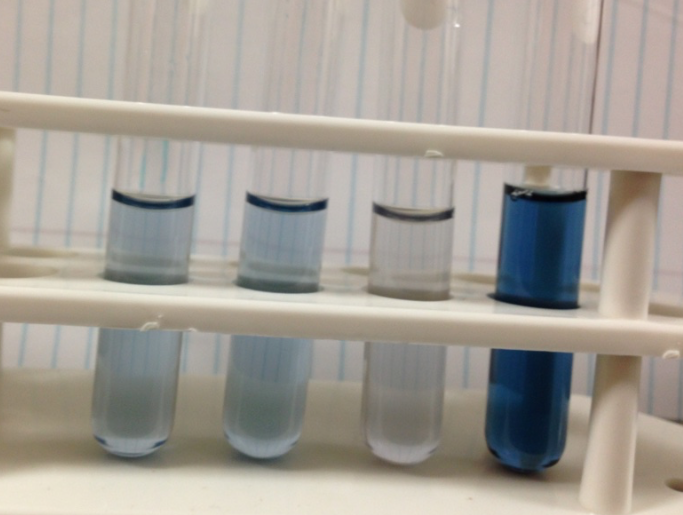
Figure 1. The image above was taken 5 hours after adding the genetically modified yeast to a starch solution. The yeast strain containing a plasmid with this composite part is the left most tube. The right most tube is the control, containing wildtype yeast and starch. Iodine, which reacts with starch to form a blue color, was added to each solution to determine the amount of starch left. The left most tube has a light blue color, indicating a small amount of starch left. The right most tube has a dark blue color, indicating a large amount of starch left. This shows that this composite part is fully functional.
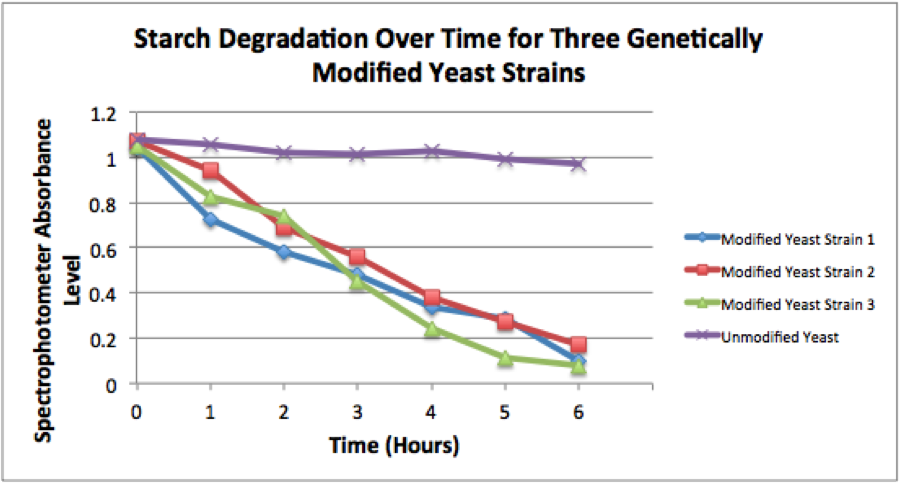
Figure 2. The image above shows a quantification of starch degradation over a period of 6 hours. A spectrophotometer was used to quantify the amount of blue color in each tube, seen in the figure above. An absorbance level of 1 equals a starch concentration of 0.25%. The yeast strain with this composite part is labeled "Modified Yeast Strain 1".
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1522
Illegal XhoI site found at 1545 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 114
Illegal AgeI site found at 336 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 650
Illegal BsaI site found at 722
| None |