Part:BBa_K1911005
eGFP
eGFP = Enhanced green fluorescent protein which can be used as a reporter gene. eGFP was used in our project to show the effects of attaching the DAS and LAA degradation tag to a target gene which is then degraded by the ClpXP system. In essence, the eGFP should be less prominent in E. coli cells which contain DAS tags and LAA tags compared to the E. coli cells that have constructs which do not have any degradation tags attached to the eGFP. eGFP can be used as a reporter gene and helps the observer easily see the coloration of E. coli cells which indicate a successful transformation.

Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 4
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 647
Usage and Biology: Characterization
TAPG tunghai 2019 - Characterization of Fluorescence intensity
The TAPG tunghai team 2019 has constructed a fusion protein by using our peptide JJ01’s back end combined with eGFP, which was induced by IPTG for expression, to ensure the accuracy of our protein expression. The fluorescent protein was employed as the identification, indicating the yield of our protein. Among of these, we have investigated four different cells in total (normal BL21, BL21 with fluorescent protein, BL21 adding IPTG, BL21 with fluorescent protein adding IPTG), and designed three strategies to ensure the accuracy.
In Figure 1. he x-axis refers to time, the y-axis refers to OD600 value. We measured the OD600 of four cells during the experiments, continually to 24 hours, and plotted the growth curve. Apparently, there is no obvious gap among these four cells.
In Figure 2. the x-axis refers to time, the y-axis refers to Fluorescence intensity. We then quantified four cells’ Fluorescence intensity, individually (Ex/Em=488/518nm), we could observe that, the curves of two cells without eGFP were not overlapping, and at the very bottom of the chart. As for the cell with eGFP but without IPTG induction, the curve remained horizontal and sended out few fluorescent. When IPTG are adding in the cell with fluorescent, we could notice that the curve has an obvious change. After adding 1 mM of IPTG, while OD600 reached to 0.8, the curve displayed a dramatic growth.
In Figure 3.the x-axis refers to time, the y-axis refers to Fluorescence intensity. We divided Figure 2. by Figure 1. and got the Fluorescence intensity of four different kind of cells per unit; then we were able to compare the productions correctly by knowing Fluorescence intensity from each cells. Because the quantity of Fluorescence protein was proportional to the target gene that was expressed. In order to know the Fluorescence intensity in each bacteria, we have to divide the numbers of bacteria by total Fluorescence intensity, which would provide us the quantity of the Fluorescence intensity per bacteria.
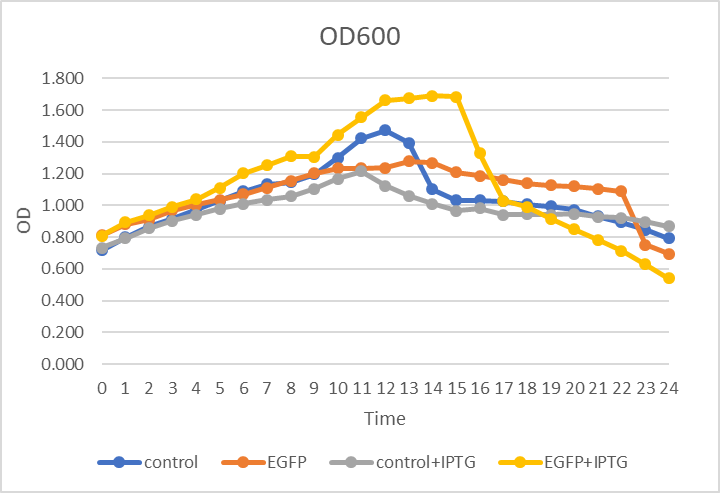
Figure.1 OD600

Figure.2 Ex/Em=488/518 nm

Figure.3 Ex/Em=488/518 nm normalized with OD600
the Condition of the Characterization
Day 1 ↓ culture E. coli BL21(DE3) carrying vector only or T7-RBS-EGFP in LB + Kan (50 ug/ml) O/N at 37°C
Day 2 ↓ measure OD600
↓ dilute to OD600 around 0.1
↓ shake at 200 rpm, 37°C until OD600 around 0.8
↓ add 1mM IPTG for protein induction
↓ measure OD600 and Ex/Em=488/518 nm every 30min for 24hr
Sequence and Features
It is tunghai_tapg igem biobrick bba_k2920001.

NYC Empire State 2022 - Recombinant Plasmid with human insulin receptor, FGF-2, NT-3
The NYC Empire State team designed a biological Trojan horse that consisted of two proteins. The first was Human Insulin Receptor Monoclonal Antibody (HIRMAb), which can interact with the HIR (Human Insulin Receptor) on endothelial cells making up the Blood-Brain Barrier to carry a cargo across the cell and into neurons. The second is a "therapeutic protein," either FGF-2 (Fibroblast Growth Factor 2) or NT-3 (Neurotrophin-3). FGF-2 binds to FGFR2 (Fibroblast Growth Factor Receptor 2), while NT-3 binds to NTRK3 (Neurotrophic Receptor Tyrosine Kinase 3).
Figure 1: Plasmid coding for FGFR2-eGFP (BBa_K4482004)

Figure 2: Plasmid coding for NTRK3-eGFP (BBa_K4482005)

Figure 3: Plasmid coding for HIR-eGFP (BBa_K4482013)
After cloning our recombinant plasmids FGFR2-eGFP, NTRK3-eGFP, and HIR-eGFP, we cut the plasmids with restriction enzymes and ran the fragments on a gel. We used Benchling to model the expected results of the experiment, and produced results that conformed to our expectations.

Figure 4: Results of running receptor sequences on a gel with restriction enzymes compared to expected results on Benchling
To test if the components of our Trojan horse could interact with their respective cell receptors, we transfected plasmids coding for the sequences of HIR, FGFR2, and NTRK3, all attached to the sequence for eGFP, into CHO-K1 cells. eGFP was attached to the C-terminus of these plasmids. After transfecting our recombinant plasmids into CHO-K1 cells, we used a fluorescent microscope to qualitatively determine that our plasmids were transfected without any problems.

Figure 5: Results of transfection with FGFR2

Figure 6: Results of transfection with NTRK3

Figure 7: Results of fluorescence microscopy with HIR
References
Cinelli RA, Ferrari A, Pellegrini V, Tyagi M, Giacca M, Beltram F. The enhanced green fluorescent protein as a tool for the analysis of protein dynamics and localization: local fluorescence study at the single-molecule level. Photochem Photobiol. 2000; 71: 771–776.
Day RN, Davidson MW. (2009) The fluorescent protein palette: tools for cellular imaging. Chem Soc Rev 38: 2887–2921
Haupts U., Maiti S., Schwille P. & Webb W. W. Dynamics of fluorescence fluctuations in green fluorescent protein observed by fluorescence correlation spectroscopy. Proceedings of the National Academy of Sciences of the United States of America 95, 13573–13578 (1998).
Kant R., Rintahaka J., Yu X., Sigvart‐Mattila P., Paulin L., Mecklin J. P., Saarela M., Palva A., and von Ossowski I.. 2014. A comparative pan‐genome perspective of niche‐adaptable cell‐surface protein phenotypes in Lactobacillus rhamnosus . PLoS ONE 9:e102762.
Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992; 111(2):229–33. doi:http://dx.doi.org/10.1016/S0167-7799(98)01184-6.
Remington S. J. (2011) Green Fluorescent Protein: A Perspective. Protein Sci. 20, 1509–1519.
Seibel N. M., Eljouni J., Nalaskowski M. M. & Hampe W. Nuclear localization of enhanced green fluorescent protein homomultimers. Analytical biochemistry 368, 95–99, doi: (2007).10.1016/j.ab.2007.05.025
Snapp, E. (2005). Design and use of fluorescent fusion proteins in cell biology. Current protocols in cell biology/editorial board, Juan S. Bonifacino ... [et al.]; CHAPTER:Unit-21.4.
Stepanenko OV, Verkhusha VV, Kuznetsova IM, Uversky VN, Turoverov KK. Fluorescent proteins as biomarkers and biosensors: Throwing color lights on molecular and cellular processes. Curr Protein Pept Sci. 2008;9:338–369.
Zhang G., Gurtu V. & Kain S. R. An enhanced green fluorescent protein allows sensitive detection of gene transfer in mammalian cells. Biochem Biophys Res Commun 227, 707–711 (1996).| None |
