Part:BBa_K1911001
pLac-ClpXP-CI
The pLac-ClpXP-CI is a transcriptional regulatory system that can work in concert with other transcriptional regulatory systems, such as p-Lambda-r=LacI (BBa_K19110000), in order to switch from one metabolic pathway to another. For example, if this part were to be inserted into E. coli cells, as evident by BBa_K1911004, the LacI repressor molecules would inhibit the pLac promoter from driving the expression of ClpXP and C.I. However, once induced with IPTG, IPTG molecules will deactivate LacI repressor molecules, thereby allowing ClpXP and C.I. to be expressed. Once expressed, the C.I. will inhibit the p-Lambda-r promoter from further synthesizing LacI repressor molecules and hence, serve as a genetic toggle switch.

Data
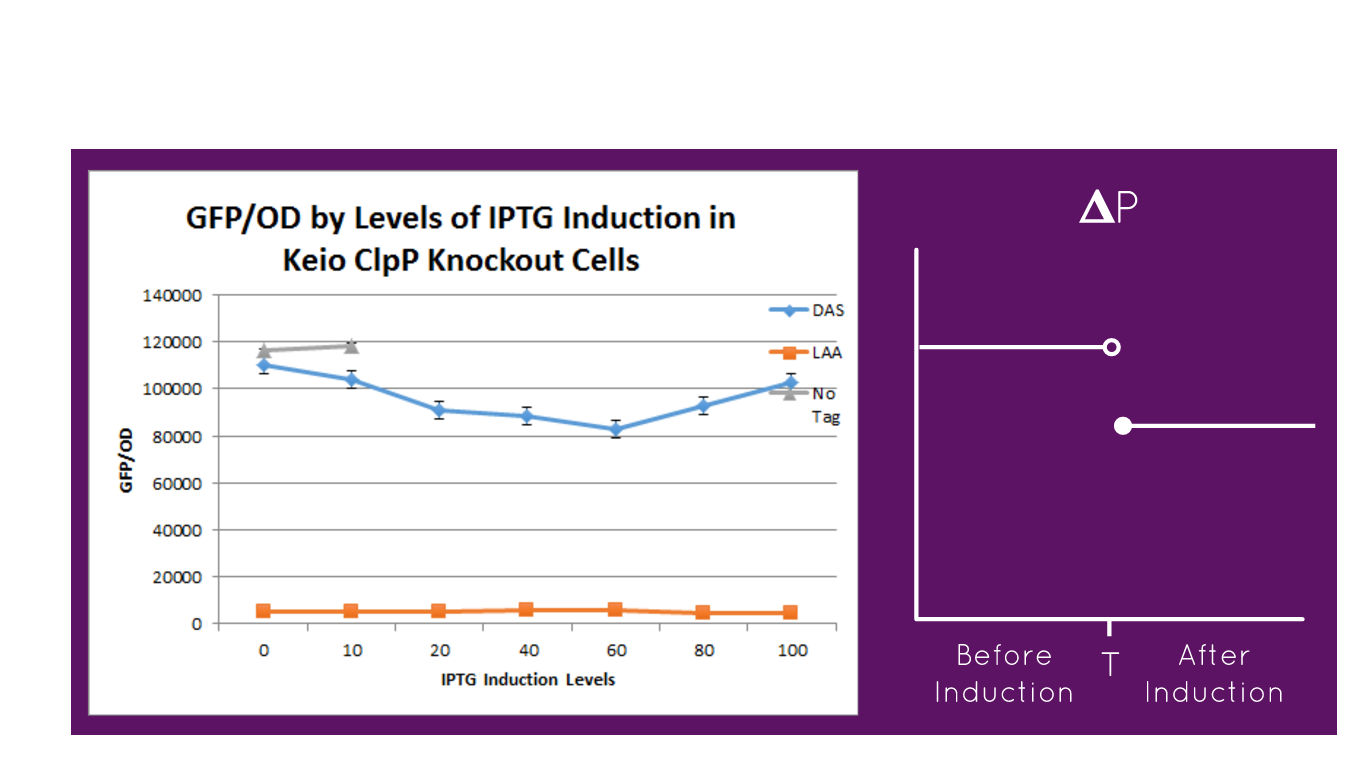
The graph above depicts the experiment in which our full construct with GFP (no tag, DAS tag, LAA tag) was grown in the Keio ClpP Knockout E. coli strains. Then, it was induced with various IPTG induction levels to allow for the expression of the ClpXP degradation system BBa_K1911001 and the GFP/OD level was observed in each of the E. coli cells. The expected results of the experiment were that the no tag would have no decrease in the GFP/OD level as IPTG increased, DAS tag would have a moderate decrease in the GFP/OD level as IPTG increased but not as much of a decrease relative to the Keio Wild Type E. coli cells experiment, and LAA tag would have stronger decrease in the GFP/OD level as IPTG increased but not as much of a decrease relative to the Keio Wild Type E. coli cells experiment. In the actual experiment, the results showed that the GFP/OD level for no tag relatively stayed the same when IPTG increased which was expected, the GFP/OD level for DAS tag decreased when IPTG increased which was expected, and the GFP/OD level for LAA tag decreased more relative to the DAS tag when IPTG increased which was also expected. The no tag data did not have as many data points as the LAA and DAS tag data because there was no growth for the E. coli colonies during the 30-80 micromolar IPTG levels. We theorize this is due to improper inoculation from the colonies. The Keio Wild Type inherently has the ClpXP system existing inside its E. coli cells.
The graph above validates our BioBrick part K1911000 p-Lambda-r=LacI because by comparing the degradation of the three different constructs (no tag, DAS tag, LAA tag) we demonstrated that the "switch system worked. As soon as the system was induced with the IPTG, the ClpXP system began to be expressed by pLac and stopped the expression the GFP, causing the GFP/OD levels to decrease. The p-Lamda-r=LacI part is shown to be working as LacI was repressing pLac, and then when the cells were induced with IPTG, the IPTG stopped LacI from repressing pLac, and the CI began to repress p-Lamda-r=LacI. The data shows that our construct is functioning properly.
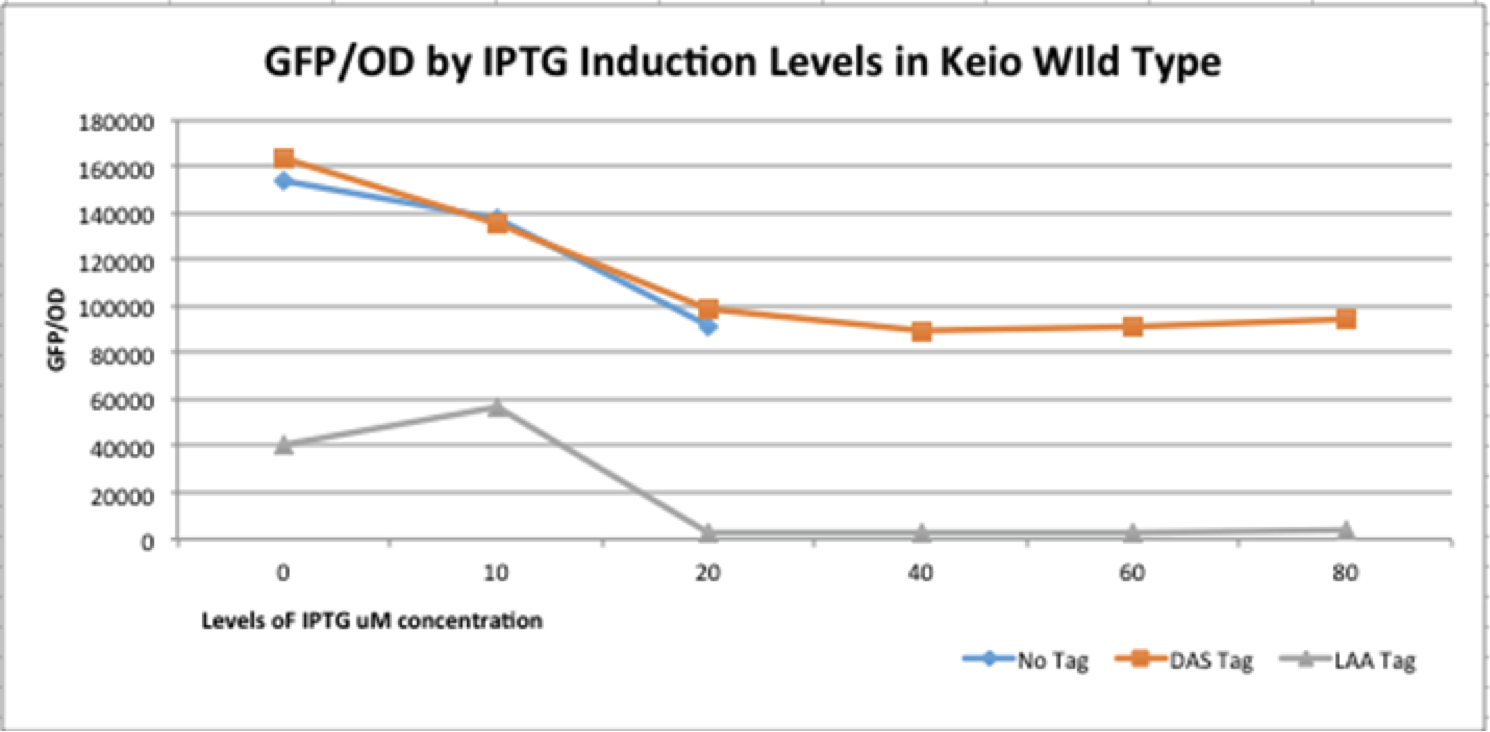
The graph above depicts the experiment in which our full construct with GFP (no tag, DAS tag, LAA tag) was grown in the Keio Wild Type E. coli strains. Then, it was induced with various IPTG induction levels to allow for the expression of the ClpXP degradation system BBa_K1911001 and the GFP/OD level was observed in each of the E. coli cells. The expected results of the experiment were that the no tag would have no decrease in the GFP/OD level as IPTG increased, DAS tag would have a moderate decrease in the GFP/OD level as IPTG increased, and LAA tag would have stronger decrease in the GFP/OD level as IPTG increased. In the actual experiment, the results showed that the GFP/OD level for no tag decreased when IPTG increased which was not expected, the GFP/OD level for DAS tag decreased when IPTG increased which was expected, and the GFP/OD level for LAA tag decreased more relative to the DAS tag when IPTG increased which was also expected. The no tag data did not have as many data points as the LAA and DAS tag data because there was no growth for the E. coli colonies during the 30-80 micromolar IPTG levels. We theorize this is due to improper inoculation from the colonies. The Keio Wild Type inherently has the ClpXP system existing inside its E. coli cells.
The graph above validates our BioBrick part K1911000 p-Lamda-r=LacI because by comparing the degradation of the three different constructs (no tag, DAS tag, LAA tag) we demonstrated that the "switch system worked. As soon as the system was induced with the IPTG, the ClpXP system began to be expressed by pLac and stopped the expression the GFP, causing the GFP/OD levels to decrease. The p-Lamda-r=LacI part is shown to be working as LacI was repressing pLac, and then when the cells were induced with IPTG, the IPTG stopped LacI from repressing pLac, and the CI began to repress p-Lamda-r=LacI. The data shows that our construct is functioning properly.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1096
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 578
Illegal AgeI site found at 1316 - 1000COMPATIBLE WITH RFC[1000]
References
Anderson LM, Yang H. DNA looping can enhance lysogenic CI transcription in phage lambda. Proc. Natl. Acad. Sci. U. S. A. 2008;105:5827–5832.
Becker N. A., Greiner A. M., Peters J. P. & Maher L. J. Bacterial promoter repression by DNA looping without protein-protein binding competition. Nucleic Acids Res. 42, 5495–5504 (2014).
Camsund D, Heidorn T, Lindblad P. Design and analysis of Lacl-repressed promoters and DNA-looping in a cyanobacterium. J Biol Eng. 2014;8:23. doi: 10.1186/1754-1611-8-4.
Kant R., Rintahaka J., Yu X., Sigvart‐Mattila P., Paulin L., Mecklin J. P., Saarela M., Palva A., and von Ossowski I.. 2014. A comparative pan‐genome perspective of niche‐adaptable cell‐surface protein phenotypes in Lactobacillus rhamnosus . PLoS ONE 9:e102762.
Khan et al. (2008) Khan SR, Gaines J, Roop II RM, Farr SK. Broad-host-range expression vectors with tightly regulated promoters and their use to examine the influence of TraR and TraM expression on Ti plasmid quorum sensing. Applied and Environmental Microbiology. 2008;74:5053–5062. doi: 10.1128/AEM.01098-08.
Lapique N., Benenson Y. Digital switching in a biosensor circuit via programmable timing of gene availability. Nat. Chem. Biol. 2014;10:1020–1027.
Lewis D, Le P, Zurla C, Finzi L, Adhya S. Multilevel autoregulation of lambda repressor protein CI by DNA looping in vitro. Proc. Natl. Acad. Sci. U. S. A. 2011;108:14807–14812.
Rogers J. K. et al. Synthetic biosensors for precise gene control and real-time monitoring of metabolites. Nucleic Acids Res. 43, 7648–7660 (2015).
Semsey S, Jauffred L, Csiszovszki Z, Erdőssy J, Stéger V, Hansen S, Krishna S. The effect of LacI autoregulation on the performance of the lactose utilization system in Escherichia coli. Nucleic Acids Res. 2013. May:gkt351.
Yang S., Sleight S. C., Sauro H. M. (2013). Rationally designed bidirectional promoter improves the evolutionary stability of synthetic genetic circuits. Nucleic Acids Res. 41, e33. 10.1093/nar/gks972
| None |
