Part:BBa_K1911000
pLamdaR-LacI
p-Lambda-r=LacI is a transcriptional regulatory system that can work in concert with other transcriptional regulatory systems, such as pLac=CI, in order to switch from one metabolic pathway to another. For example, if this part were to be inserted into E. coli cells and induced with IPTG, the IPTG would bind to the LacI repressor, thereby allowing the system to switch from one pathway to the next. Once expressed, the second metabolic pathway will produce the C.I. repressor molecules, which will inhibit the p-Lambda-r promoter from further sythesizing LacI molecules and the gene of interest. This part was created in order for teams to be able to easily place their genes of interest in this "switch" system. They would combine their gene of interest with the pLamdaR-LacI part and then proceed to combine it to the rest of the inducible system.

Data
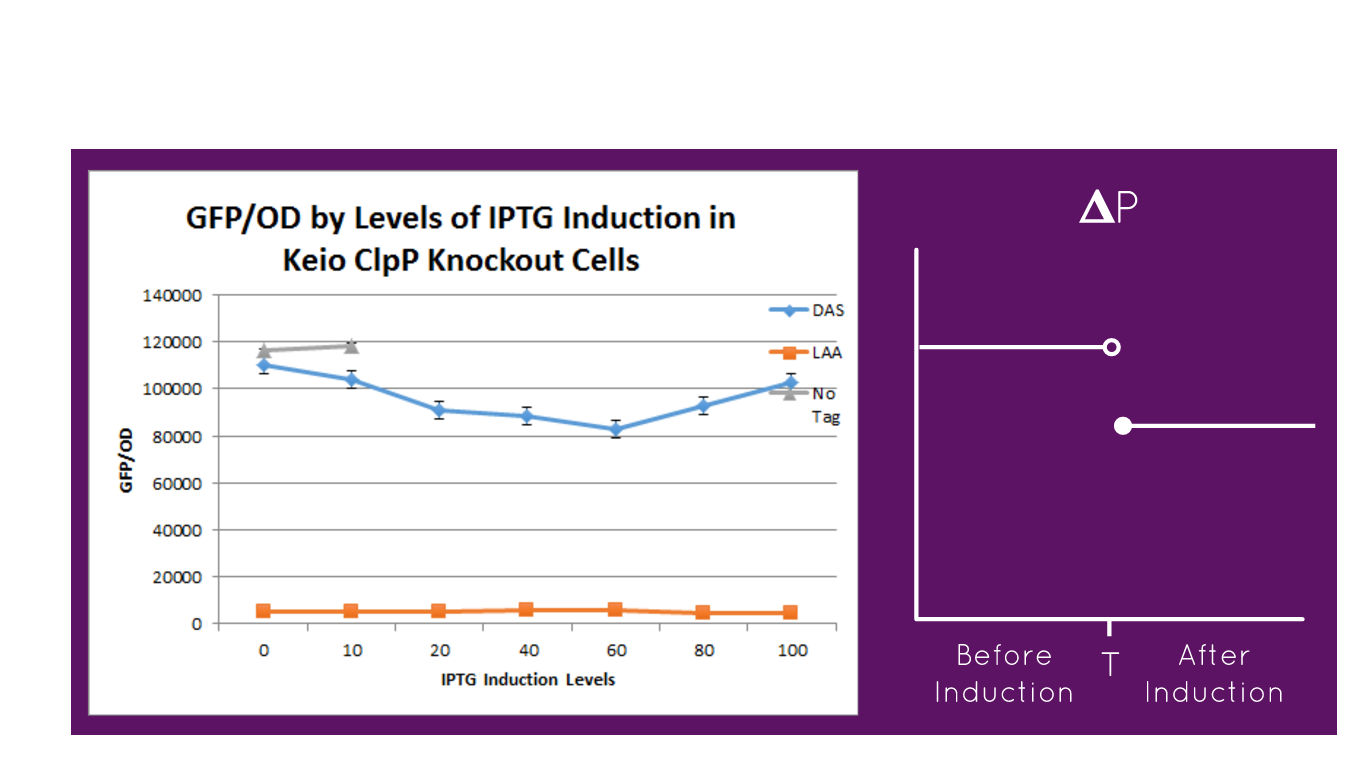
The graph above depicts the experiment in which our full construct with GFP (no tag, DAS tag, LAA tag) was grown in the Keio ClpP Knockout E. coli strains. Then, it was induced with various IPTG induction levels to allow for the expression of the ClpXP degradation system BBa_K1911001 and the GFP/OD level was observed in each of the E. coli cells. The expected results of the experiment were that the no tag would have no decrease in the GFP/OD level as IPTG increased, DAS tag would have a moderate decrease in the GFP/OD level as IPTG increased but not as much of a decrease relative to the Keio Wild Type E. coli cells experiment, and LAA tag would have stronger decrease in the GFP/OD level as IPTG increased but not as much of a decrease relative to the Keio Wild Type E. coli cells experiment. In the actual experiment, the results showed that the GFP/OD level for no tag relatively stayed the same when IPTG increased which was expected, the GFP/OD level for DAS tag decreased when IPTG increased which was expected, and the GFP/OD level for LAA tag decreased more relative to the DAS tag when IPTG increased which was also expected. The no tag data did not have as many data points as the LAA and DAS tag data because there was no growth for the E. coli colonies during the 30-80 micromolar IPTG levels. We theorize this is due to improper inoculation from the colonies. The Keio Wild Type inherently has the ClpXP system existing inside its E. coli cells.
The graph above validates our BioBrick part K1911000 p-Lambda-r=LacI because by comparing the degradation of the three different constructs (no tag, DAS tag, LAA tag) we demonstrated that the "switch system worked. As soon as the system was induced with the IPTG, the ClpXP system began to be expressed by pLac and stopped the expression the GFP, causing the GFP/OD levels to decrease. The p-Lamda-r=LacI part is shown to be working as LacI was repressing pLac, and then when the cells were induced with IPTG, the IPTG stopped LacI from repressing pLac, and the CI began to repress p-Lamda-r=LacI. The data shows that our construct is functioning properly.
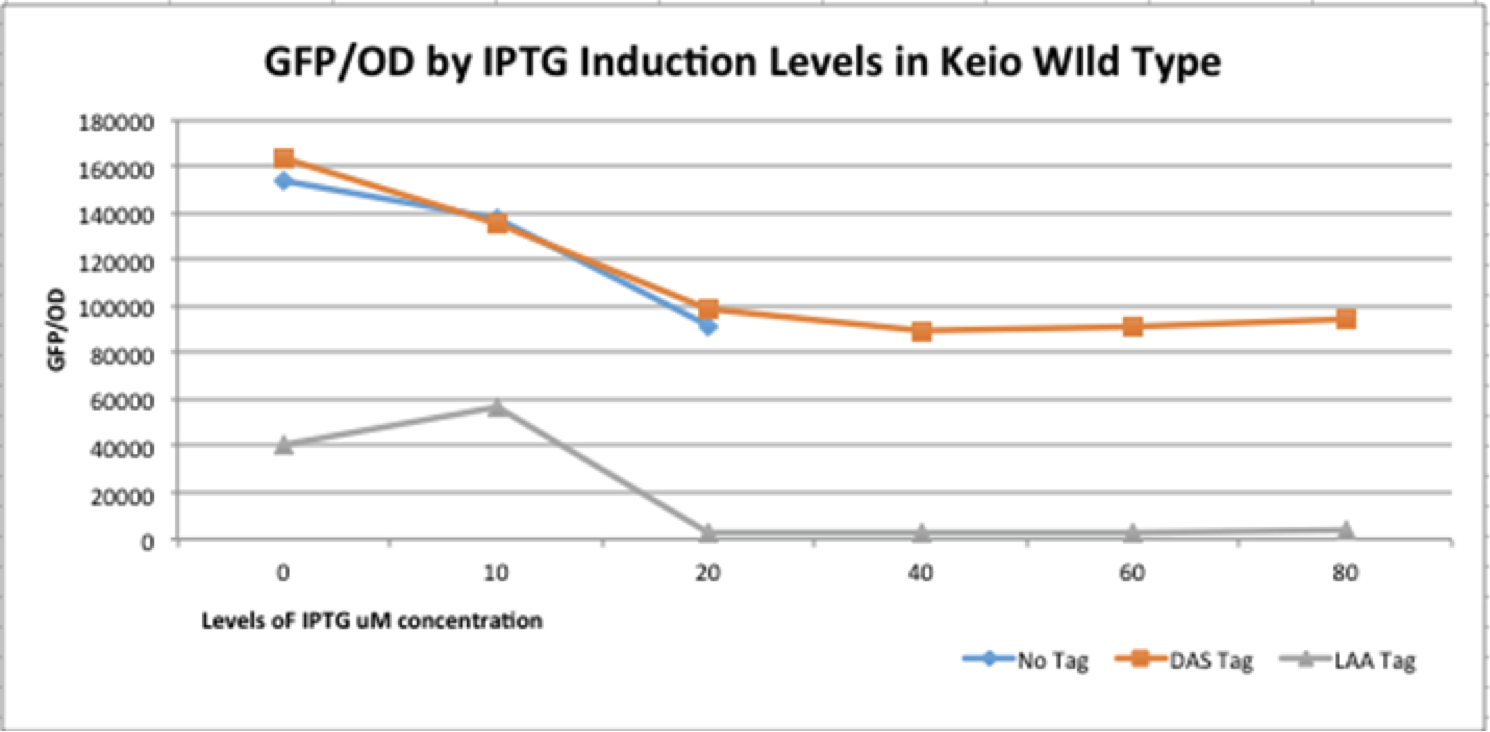
The graph above depicts the experiment in which our full construct with GFP (no tag, DAS tag, LAA tag) was grown in the Keio Wild Type E. coli strains. Then, it was induced with various IPTG induction levels to allow for the expression of the ClpXP degradation system BBa_K1911001 and the GFP/OD level was observed in each of the E. coli cells. The expected results of the experiment were that the no tag would have no decrease in the GFP/OD level as IPTG increased, DAS tag would have a moderate decrease in the GFP/OD level as IPTG increased, and LAA tag would have stronger decrease in the GFP/OD level as IPTG increased. In the actual experiment, the results showed that the GFP/OD level for no tag decreased when IPTG increased which was not expected, the GFP/OD level for DAS tag decreased when IPTG increased which was expected, and the GFP/OD level for LAA tag decreased more relative to the DAS tag when IPTG increased which was also expected. The no tag data did not have as many data points as the LAA and DAS tag data because there was no growth for the E. coli colonies during the 30-80 micromolar IPTG levels. We theorize this is due to improper inoculation from the colonies. The Keio Wild Type inherently has the ClpXP system existing inside its E. coli cells.
The graph above validates our BioBrick part K1911000 p-Lamda-r=LacI because by comparing the degradation of the three different constructs (no tag, DAS tag, LAA tag) we demonstrated that the "switch system worked. As soon as the system was induced with the IPTG, the ClpXP system began to be expressed by pLac and stopped the expression the GFP, causing the GFP/OD levels to decrease. The p-Lamda-r=LacI part is shown to be working as LacI was repressing pLac, and then when the cells were induced with IPTG, the IPTG stopped LacI from repressing pLac, and the CI began to repress p-Lamda-r=LacI. The data shows that our construct is functioning properly.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1221
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
References
Atsumi S., and Little J. W.. 2004. Regulatory circuit design and evolution using phage λ . Genes Dev.18:2086–2094.
Chia N, Golding I, Goldenfeld N. Lambda-prophage induction modeled as a cooperative failure mode of lytic repression. Phys Rev E Stat Nonlin Soft Matter Phys. 2009;80:030901.
Dodd IB, Egan JB. Action at a distance in CI repressor regulation of the bacteriophage 186 genetic switch. Mol Microbiol. 2002;45: 697–710. doi: 10.1046/j.1365-2958.2002.03038.x
Dodd IB, Perkins AJ, Tsemitsidis D, Egan JB. Octamerization of lambda CI repressor is needed for effective repression of P(RM) and efficient switching from lysogeny. Genes. Dev. 2001;15:3013–3022.
Dodd I.B., Shearwin K.E., Perkins A.J., Burr T., Hochschild A., Egan J.B. Cooperativity in long-range gene regulation by the lambda CI repressor. Genes Dev. 2004;18:344–354. doi: 10.1101/gad.1167904.
Fong RS, Woody S, Gussin GN. Modulation of P(RM) activity by the lambda PR promoter in both the presence and absence of repressor. J Mol Biol. 1993;232:792–804.
Hayes S, Horbay MA, Hayes C (2012) A CI-Independent Form of Replicative Inhibition: Turn Off of Early Replication of Bacteriophage Lambda. PLoS ONE 7(5): e36498. doi:10.1371/journal.pone.0036498
Pastrana, R., & Brammar, W. J. (1976, January 15). Control of cI gene expression in bacteriophage λimm434, studied in an immunity/trp fusion made in vitro. Molecular and General Genetics MGG, 146(2), 191-198. doi:10.1007/BF00268088
Reichardt L., and Kaiser A. D.. 1971. Control of lambda repressor synthesis. Proc. Natl. Acad. Sci. USA 68:2185–2189.
Santillán M., Mackey M. C. (2004). Why the lysogenic state of phage λ is so stable: a mathematical modeling approach. Biophys. J. 86, 75–84. 10.1016/S0006-3495(04)74085-0
Schubert R. A., Dodd I. B., Egan J. B., and Shearwin K. E.. 2013. Cro's role in the CI‐Cro bistable switch is critical for λ's transition from lyogeny to lytic development. Genes Dev. 21:2461–2472.
Svenningsen SL, Costantino N, Court DL, Adhya S. On the role of Cro in lambda prophage induction. Proc Natl Acad Sci U S A. 2005;102(12):4465–9. doi: 10.1073/pnas.0409839102 ; PubMed Central PMCID: PMCPMC555511.
| None |
