Part:BBa_K1887001
His
The His operon is required for the histidine biosynthesis in L. lactis,the His operon contains several genes.Here we chose one segment of the His operon for device knocked into the genome of L.lactis. In other words, the His segment is used as the flanking sequences for homologous recombination.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Molecular verification
We used PCR and restriction enzyme digestion to verify the His part on pSB1C3.
Background
Currently, most of the devices are introduced into L. lactis by plasmids. Various vectors containing constitutive or inductive promoters have been developed, such as pMG36e, pNZ8148. However, the devices introduced by plasmids had two disadvantages. First, most plasmids have selection markers (genes) for antibiotic resistance, which is forbidden to be used in food or digestive tract of human and animals directly. Second, the plasmids were unstable in cytoplasm in the absence of selection pressure.
The alternative choice is knocking the devices into the genomes of L. lactis. Double cross over recombination using non-replicative or conditional replicative plasmids is the widely used strategy. For instance, using temperature sensitive plasmid, at restrictive temperature, single-crossover integration of devices can be selected by certain antibiotics. Under lower temperature and in the absence of antibiotics, the second cross-over is occurred at a very low rate and the desired device knocked-in strains can be obtained. However, to get desired device knocked-in strains, antibiotic resistance of clones was checked, and only a few clones were antibiotic susceptible. The antibiotic susceptible clones were then verified by PCR and less than half of them were the desired device knocked-in strains, the others were the parental strains.
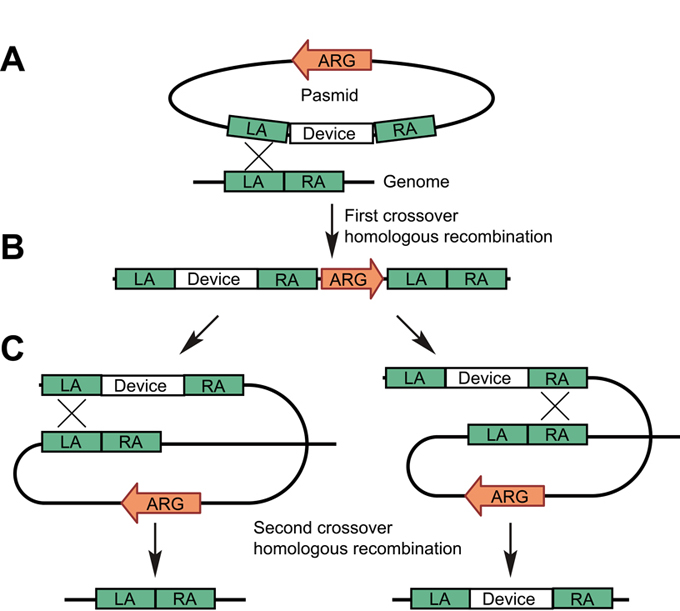
Schematic of device integration. Abbreviations: LA, left arm for integration; RA, right arm for integration; ARG, antibiotic resistant gene.
Design
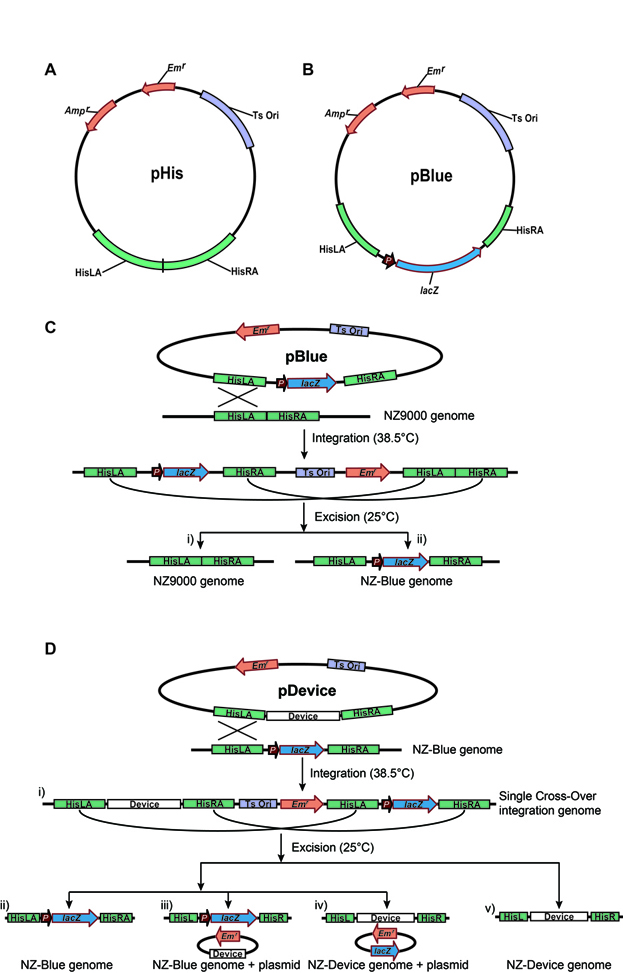
To get device knocked-in recombinant L. lactis without antibiotic resistant genes and make this process easy and highly efficient, we try to establish a visual selection system based on the insoluble blue compounds generated by the lacZ gene encoded β-galactosidase catalyzed X-gal (also abbreviated BCIG for 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) hydrolysis. To build this system, we chose the His locus as the targeting locus. The His locus is required for the synthesis of histidine, an essential amino acid for L. lactis growth. The recombinant L. lactis strains can only be survived with the supplementation of histidine in the culture but cannot survive without histidine. The His fragment was introduced into a temperature-sensitive conditional replicative shuttle plasmid and we named this plasmid as pHis. Then we placed the lacZ gene driven by the PnisZ promoter into the His locus and we name this plasmid as pBlue. The next step is to integrate the lacZ gene into the genome of NZ9000 to yield the NZ-Blue recombinant strain. The lacZ gene encoded β-galactosidase can hydrolyze the X-gal from colorless substance to insoluble blue compounds, thus NZ-blue strain forms blue colonies on X-gal plate supplemented with nisin. Finally, the device carrying plasmid based on pHis (pDevice) will be introduced to NZ-Blue to replace the lacZ gene and the desired device knocked-in strains will be white when cultured with X-gal and supplemented with nisin.
The markerless integration visual selection system for L. lactis. A). Map of the integration plasmid pHis; B). Map of the integration plasmid pBlue; C). Schematic of the construction of NZ-Blue; D). Schematic of high-efficiency device integration selection. Abbreviations: His, integration fragment from L. lactis; HisLA, left arm of His; HisRA, right arm of His; Ts Ori, temperature-sensitive replicon from pCrePA2; Emr, erythromycin resistant gene; Ampr, ampicillin resistant gene; P, PnisZ, the promoter of lacZ; lacZ, β-galactosidase from Lactobacillus acidophilus.
Results
The temperature-sensitive conditional replicative plasmid pHis was used to introduce the ‘PnisZ:: lacZ::Terminator’ fragments into the NZ9000 chromosome of L. lactis. The constructed plasmid pBlue was transformed into NZ9000 by electroporation. To obtain NZ-Blue via homologous sequences (HisLA or HisRA), the NZ9000-pBlue strains were grown in M17GS (M17 broth with 0.55 % (w/v) sucrose and 0.5 % (w/v) glucose) containing erythromycin and incubated at 38.5 °C overnight to integrate the pBlue DNA sequence into the genome of NZ9000 through single cross over. Then, the cultures were diluted in M17GS medium without erythromycin and incubated at 28 °C to promote the double cross over and excision of the antibiotic resistance genes. The subcultures were diluted and plated on M17GS with X-Gal and nisin. Blue colonies obtained by serial plating were screened for erythromycin susceptibility. Colonies in which lacZ gene integration had occurred will appear as blue on X-gal plate supplemented with nisin and erythromycin susceptible. The erythromycin susceptible and blue colonies selected from thousands of colonies were further subject to PCR verification using the extracted genomic DNA as PCR template. DNA sequencing was also used to confirm that no genetic mutations were introduced during the experiment process. After this difficult and tedious task, we successfully got the lacZ integration strain and we name it NZ-Blue.
Confirmation analysis of the constructed NZ-Blue strain. A). Schematic of genomes of NZ9000 and NZ-Blue. Abbreviations: LO, RO, LF, LR, primers used in Multiple-PCR analysis; HisLA, left arm of His; HisRA, right arm of His; PnisZ, promoter induced by nisin; lacZ, β-galactosidase from Lactobacillus acidophilus; Ter, terminator. B). Multiple-PCR analysis of the NZ-Blue and NZ9000. The primer combinations used in PCR are presented upon the lanes. Genomic DNA used in PCR are presented upon the lines. C). Colony of NZ-Blue. The 1 kb DNA ladder marker is shown to the left (M).
References
[1] Delorme, C., Ehrlich, S.D., and Renault, P. (1992). Histidine biosynthesis genes in Lactococcus lactis subsp. lactis. J Bacteriol 174, 6571-6579.
[2] Simoes-Barbosa, A., Abreu, H., Silva Neto, A., Gruss, A., and Langella, P. (2004). A food-grade delivery system for Lactococcus lactis and evaluation of inducible gene expression. Appl Microbiol Biotechnol 65, 61-67.
| None |