Part:BBa_K1680015
pRS315 yeast shuttle vector with Biobrick MCS
This part is a biobricked shuttle vectors for Saccharomyces cerevisiae. It is part of a vector series with different auxotrophy markers (Part:BBa_K1680014, Part:BBa_K1680016). The vector was made compatible with the RFC10 standard and also contains a RFC10 cloning site. This part is derived from the wild type pRS315 vector Part:BBa_K950010.
- Request concerning submission for plasmid backbone was sent to HQ on 09/17
Further - not biobrick compatible - versions of this vector with different inserts can also be found in the registry: Part:BBa_K106006, Part:BBa_K106014, Part:BBa_K106670, Part:BBa_K106672, Part:BBa_K106693, Part:BBa_K106697, Part:BBa_K106698
This part encodes for an ampicillin resistance gene and replicates to high-copy numbers in E. coli. Furthermore, the CEN6/ARS4 (Chromosome VI centromere/Autonomously Replicating Sequence 4) cassette ensures that the plasmid stays at a low copy number in yeast, because they are treated as pseudo chromosomes. The vector also encodes for a LEU2 gene, which is used as an auxotrophy marker in yeast.
This part is mutagenized version of the wild type pRS315 vector (Sikorski 1989).
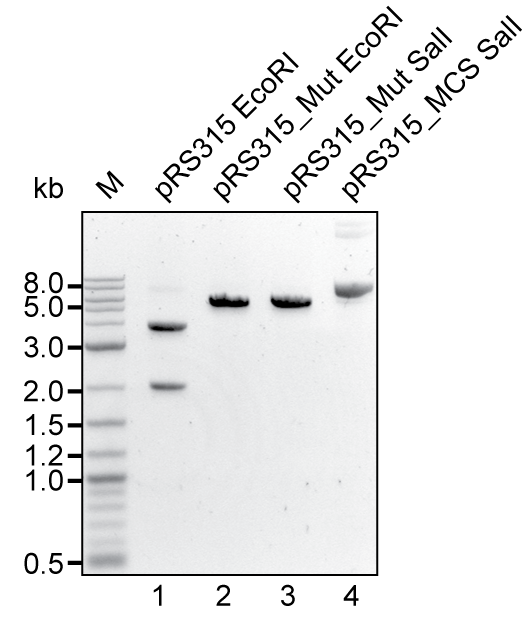
A EcoRI restriction site in the backbone was removed by mutagenesis PCR (see lane 1 & 2 in figure). Exchange of the wild type MCS for RFC10 compatible MCS could be shown by restriction with SalI restriction: The enzyme linearizes the vector with old MCS (lane 3) but does not cut the vector with the new MCS, as indicated by presence of supercoiled DNA (lane 4).
Agarose Gel of different Versions of the pRS313 vector. Lane 1: wild type vector, cut with EcoRI; lane 2: mutagenised vector cut with EcoRI; lane 3: mutagenised vector cut with SalI; lane 4: vector with RFC10 MCS cut with SalI.
Sequence and Features
This part is a RFC10 compatible shuttle vector for yeast.
- 10INCOMPATIBLE WITH RFC[10]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 3171
Illegal XbaI site found at 3156
Illegal SpeI site found at 3148
Illegal PstI site found at 3134 - 12INCOMPATIBLE WITH RFC[12]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 3171
Illegal SpeI site found at 3148
Illegal PstI site found at 3134
Illegal NotI site found at 3139
Illegal NotI site found at 3163 - 21INCOMPATIBLE WITH RFC[21]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 3171
Illegal XhoI site found at 2410 - 23INCOMPATIBLE WITH RFC[23]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 3171
Illegal XbaI site found at 3156
Illegal SpeI site found at 3148
Illegal PstI site found at 3134 - 25INCOMPATIBLE WITH RFC[25]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 3171
Illegal XbaI site found at 3156
Illegal SpeI site found at 3148
Illegal PstI site found at 3134
Illegal NgoMIV site found at 2799
Illegal AgeI site found at 1503 - 1000INCOMPATIBLE WITH RFC[1000]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal BsaI.rc site found at 4545
Illegal SapI site found at 3462
Illegal SapI site found at 5566
//dna
//plasmid
//plasmidbackbone
//plasmidbackbone/copynumber/low
| None |