Part:BBa_K165096
Zif268-HIV bs + MET25+ Gli1 repressor (CFPx2 tagged) on pRS304*
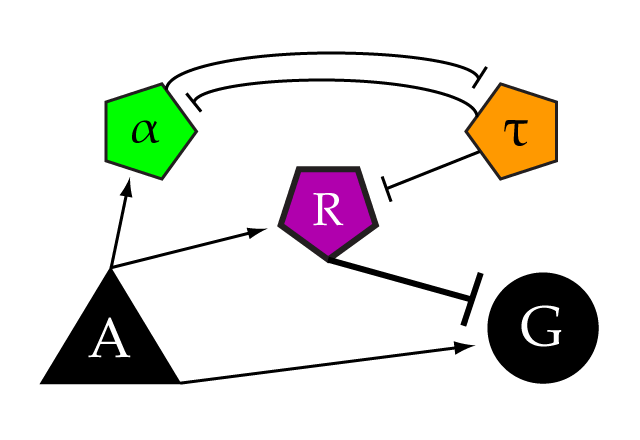
This construct is an example of the Tau element of our genetic limiter network. In this case, the pMET25 sets the threshold of limitation, allowing us to tune the threshold by varying the concentration of methionine in the media the cells are grown in. We constructed the Gli1 repressor in this devise with CFPx2 instead of mCherry, allowing us to distinguish it from the other repressors in the network. It is placed in a mutually inhibitory relationship with repressor alpha, in this case the Zif-HIV repressor. When the network is subthreshold, this tau repressor is expressed, and represses R, turning off its repression of the gene of interest.
Usage and Biology
To build a (heretofore untested) limiter, this part should be in a yeast strain with the following constructs:
A) Part:BBa_K165080 pGAL1 + Untagged LexA activator on pRS305
G) Part:BBa_K165078 LexA activable reporter on pRS306
Into mating type a. Knock out the Gal2 gene for a tunable level of induction.
R) Part:BBa_K165090 Gli1 bs + LexA bs + mCYC + LexA repressor (mCherryx2 tagged) on pRS306
Alpha) Part:BBa_K165095 Gli1 bs + LexA bs + mCYC + Zif268-HIV repressor (mCherryx2 tagged) on pRS303
Tau) Part:BBa_K165096 Zif268-HIV bs + MET25+ Gli1 repressor (CFPx2 tagged) on pRS304*
Into mating type alpha. Knock out the Gal2 gene for a tunable level of induction.
Mate the two types, select on SD-His-Trp-Leu-Ura
Inputs: Set the threshold by tuning [Met] between 0 and 500 uM Set the level of induction (A) with Galactose between 0-3%
Outputs expected:
Subthreshold [A]: CFP in the nucleus from Tau, YFP in the cytosol proportionate to subthreshold induction from G being activated by A and not repressed by R.
Superthreshold [A]: mCherry in the nucleus from Alpha and R induction. YFP in cytosol levels out at threshold limit despite rising induction.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 2224
Illegal XhoI site found at 1
Illegal XhoI site found at 41 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 3990
Illegal AgeI site found at 4740 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 1395
Illegal SapI.rc site found at 1988
//function/regulation/transcriptional
| None |

 1 Registry Star
1 Registry Star