Part:BBa_K1547001
PSAD GFP-NLS cassette
PSAD promoter and terminator sequences flanking a chlamydomonas reinhardtii codon optimized GFP, with a 3' NLS signal.
This cassette contains a synthetic codon-optimized for chlamydomonas reinhardtii GFP reporter [1], which was supplied by chlamycollection.org . More information on this codon-optimized GFP can be found here: http://chlamycollection.org/wp-content/uploads/2012/09/pCrGFP.pdf .
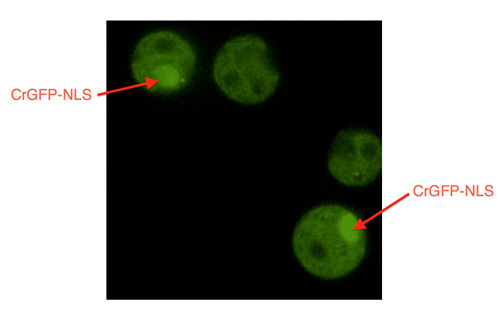
Our GFP-NLS cassette, containing a strong PSAD promoter [2] and its respective terminator, was introduced into chlamydomonas reinhardtii (UVM1) by electroporation. Characterization was carried out after transformants were then grown in antibiotic selection media in 12:12 light/dark cycles, on a shaker, for several days. Fluorescent microscopy was used to examine the presence localization the GFP. Our preliminary images produced clear images of bright green areas in the transformant cells, whereas the wildtype cells lacked any bright areas. Moreover, the GFP appeared to be localized to one specific region in the cells. We can therefore conclude that our GFP-NLS construct produces a GFP that localizes to a specific location in chlamydomonas reinhardtii cells. Further DNA staining techniques can be used to verify that this region is the location of the nucleus, and further characterize our part in that manner. Images can be seen on Concordia 2014's wiki: http://2014.igem.org/Team:Concordia/Notebook . Furthermore, we noted that the intensity of our PSAD-GFP-NLS sample appeared much higher than that of our GFP construct controlled by an HSP70/RBCS2 fusion promoter.
Transformant c. reinhardtii cells containing GFP, localized to a specific region:
Wildtype c. reinhardtii cells
1. Fuhrmann et al. (1999) Plant J. 19, 353-361
2. Harris, Elizabeth H. (2009). The Chlamydomonas Sourcebook (2nd Edition) Volume 1: Introduction to Chlamydomonas and Laboratory Use. Durham, North Carolina. Elsevier.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 1570
Illegal AgeI site found at 916 - 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 1703
| None |