Part:BBa_K1453003
pBluescript II KS(+)_3_copy
Vector
We used pBluescript II KS(+) as our origin vector and then transformed it into the Connector of our desire.
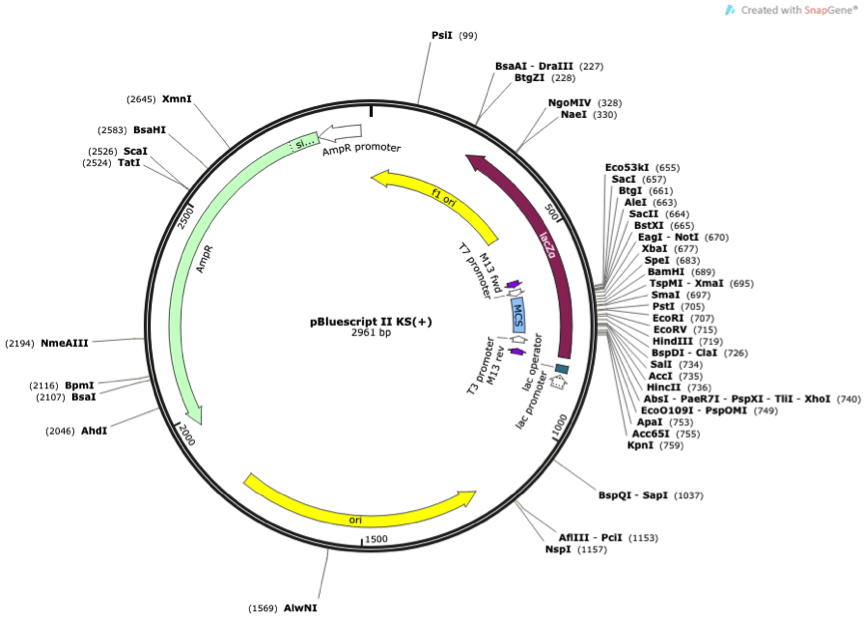
Figure 1 Vector map of pBluescript II KS(+)
Connector
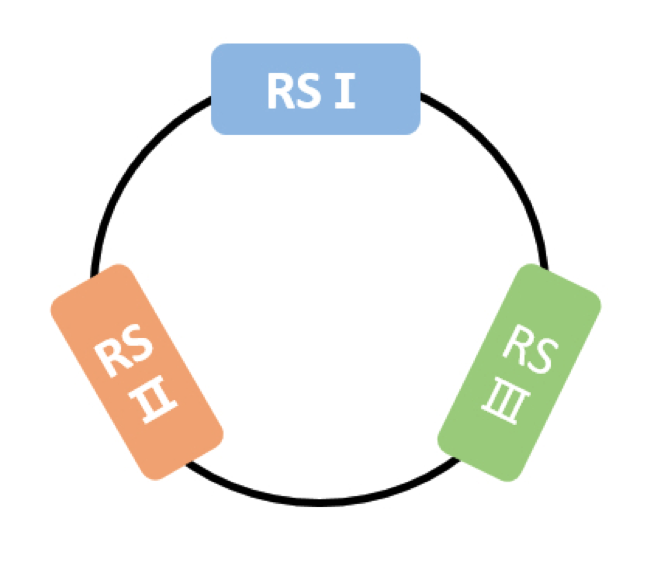
The so-called Connector is plasmid designed to bind with several different Connectees(iGEM12_SJTU_BioX_Shanghai BBa_K771000). Connectees are fusion proteins who carry out functional enzymes and have TAL(Transactivator-like effectors) at one end that can bind to certain DNA sequences and you can design your own Connectees with different enzymes. For pBluescript II KS(+), we have selected three 14-nucleotide-long sequences called RSⅠ, RSⅡ and RSⅢ as the recognition sequences(RS). The recognition sequences must start with a T and end with a T. RS Ⅰ: TTCGATATCAAGCT RS Ⅱ: TGTGACTGGTGAGT RS Ⅲ: TTTGGTCATGAGAT ( RS Ⅰ contains partial restriction enzyme cutting site of EcoRI and EcoRV ) ( RS Ⅱ contains partial restriction enzyme cutting site of ScaI )
Figure 2 A Connector binds with three different Connectees
Maximization
We are trying to combine as many enzymes as possible because more enzymes’ combination can produce more complicated reaction chains. So our purpose is to figure out the maximum number of Connectees that binding to a Connector. So we intended to add more RS on the Connectors. On the one hand, more RSs mean we can bind more kinds of enzymes on one Connector. On the other hand ,we doubt the binding efficiency between Connectors and Connectees so more RSs can improve the possibility of Connectees binding to Connectors.The corresponding Connectors are pBluescript II KS(+)_3_copy and pBluescript II KS(+)_5_copy.
pBluescript II KS(+)_3_copy
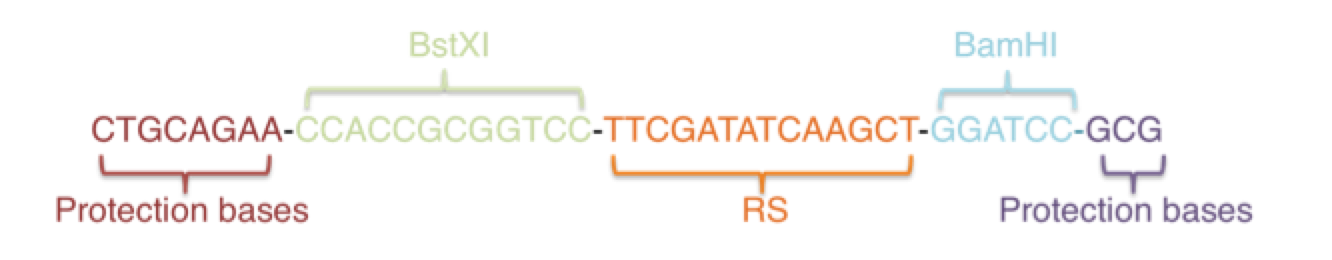
In order to add 3 copies of RS on Connectors, we firstly use restriction enzyme BstXI and BamHI to make a nick and replace the original sequence with one RS. Secondly, in the same way, we use restriction enzyme SalI and KpnI to add another RS. The RS sequence is TTCGATATCAAGCT.
Here we have designed the new short sequence:
This is the what we get in the end:
CT...CTCCACCGCGGTGGTTCGATATCAAGCTGGATCCCCCGGGCTGCAGGAAT TCGATATCAAGCTTATCGATACCGTCGACTTCGATATCAAGCTGGTACCCA….AC
pBluescript II KS(+)_3_copy can be used along with our Connectees(iGEM12_SJTU_BioX_Shanghai BBa_K771000) to achieve your certain pathway design.
Figure 3 The structure of pBluescript II KS(+)_3_copy
Usage and Biology
If you want to use our Connector, there is a list for possible TAL recognition sequences on pBluescript II KS(+). You can choose any combination of following sequences to design your own Connectees.
'TAAAAAGGCCGCGT', 'TAAAAATGAAGTTT', 'TAAATCAAAAGAAT', 'TAAATCAGCTCATT', 'TAAATCGGAACCCT', 'TAAATTGTAAGCGT', 'TAACGCCAGGGTTT', 'TAACTCACATTAAT', 'TAAGATGCTTTTCT', 'TAAGCGTTAATATT', 'TAAGGGATTTTGGT', 'TAATACGACTCACT', 'TAATCAAGTTTTTT', 'TACCAATGCTTAAT', 'TACCCAGCTTTTGT', 'TACCGGATACCTGT', 'TACCTGTCCGCCTT', 'TACTCTTCCTTTTT', 'TACTGTCATGCCAT', 'TAGAACTAGTGGAT','TAGAAGGACAGTAT', 'TAGAGTAAGTAGTT', 'TAGATAACTACGAT', 'TAGCAGAGCGAGGT', 'TAGCGGTGGTTTTT', 'TAGCTGTTTCCTGT', 'TAGGGTTGAGTGTT', 'TAGGTATCTCAGTT', 'TAGTGAGGGTTAAT', 'TATATGAGTAAACT', 'TATCACTCATGGTT', 'TATCCGCTCACAAT', 'TATCCGGTAACTAT', 'TATCTCAGCGATCT', 'TATCTCAGTTCGGT', 'TATTATTGAAGCAT', 'TATTGGGCGCTCTT', 'TATTTGAATGTATT', 'TATTTTGTTAAAAT', 'TCAAAAAGGATCTT', 'TCAAAGGCGGTAAT', 'TCAACCAAGTCATT', 'TCAAGAAGATCCTT', 'TCAAGCTTATCGAT', 'TCAATCTAAAGTAT', 'TCACCAGCGTTTCT', 'TCACCTAGATCCTT', 'TCACGCTCGTCGTT', 'TCACGTTAAGGGAT', 'TCACTCATGGTTAT', 'TCACTGCCCGCTTT', TCAGCCCGACCGCT', 'TCAGCGATCTGTCT', 'TCAGCTCATTTTTT', 'TCAGTGAGGCACCT', 'TCATACTCTTCCTT', 'TCATAGCTCACGCT', 'TCATGGTCATAGCT', 'TCATTGGAAAACGT', 'TCCAGTCTATTAAT', 'TCCATCCAGTCTAT', 'TCCCATTCGCCATT', 'TCCCCCTGGAAGCT', 'TCCGCTTCCTCGCT', 'TCCGTAAGATGCTT', 'TCCTGCAACTTTAT', 'TCCTGTGTGAAATT', 'TCCTTTGATCTTTT', 'TCCTTTTTCAATAT', 'TCGATATCAAGCTT', 'TCGCCATTCAGGCT','TCGCGTTAAATTTT','TCGCTGCGCTCGGT','TCGGAAAAAGAGTT','TCGGCAAAATCCCT','TCGGGGCGAAAACT','TCGGTCGTTCGGCT', 'TCGGTGTAGGTCGT', 'TCGTCGTTTGGTAT', 'TCGTGCACCCAACT', 'TCGTGCGCTCTCCT', 'TCGTGTAGATAACT', 'TCGTTGTCAGAAGT', 'TCTAAAGTATATAT', 'TCTATCAGGGCGAT', 'TCTATTTCGTTCAT', 'TCTCAGCGATCTGT', 'TCTCAGTTCGGTGT', 'TCTCATGAGCGGAT', 'TCTCTTACTGTCAT', 'TCTGAGAATAGTGT', 'TCTGTCTATTTCGT', 'TCTTCAGCATCTTT', 'TGAACCATCACCCT', 'TGAAGTGGTGGCCT', 'TGAATACTCATACT', 'TGACAGTTACCAAT','TGACTCCCCGTCGT', 'TGACTCGCTGCGCT','TGAGAATAGTGTAT','TGAGCGCGCGTAAT','TGAGGCACCTATCT','TGAGTAAACTTGGT','TGAGTGAGCTAACT','TGATCCCCCATGTT', 'TGCAAAAAAGCGGT','TGCAAGCAGCAGAT','TGCACCCAACTGAT','TGCAGGAATTCGAT','TGCATAATTCTCTT','TGCCCGGCGTCAAT','TGCCGTAAAGCACT', 'TGCGCAACGTTGTT','TGCGCGCTTGGCGT','TGCGCTCGGTCGTT','TGCGCTCTCCTGTT',TGCGGCGACCGAGT','TGCTACAGAGTTCT','TGCTGAAGCCAGTT', 'TGCTGCAAGGCGAT','TGGAAAACGTTCTT','TGGCAGCACTGCAT','TGGCAGCAGCCACT', 'TGGCGCTTTCTCAT', 'TGGCGTTTTTCCAT', 'TGGTAGCGGTGGTT','TGGTAGCTCTTGAT', 'TGGTATGGCTTCAT', 'TGGTCATAGCTGTT', 'TGGTCCTGCAACTT', 'TGGTTTTTTTGTTT', 'TGTAACCCACTCGT', 'TGTCAGAAGTAAGT', 'TGTGAAATTGTTAT', 'TGTGACTGGTGAGT', 'TGTGTGAAATTGTT', 'TGTTGAATACTCAT', 'TGTTGAGATCCAGT', 'TGTTGTTCCAGTTT', 'TTAAAAATGAAGTT', 'TTAAAAGTGCTCAT', 'TTAAAATTCGCGTT', 'TTAAATCAGCTCAT', 'TTAATTGCGCGCTT', 'TTAGCTCCTTCGGT', 'TTATCAAAAAGGAT', 'TTATCACTCATGGT', 'TTATCAGGGTTATT', 'TTATCCGCCTCCAT', 'TTATTGAAGCATTT', 'TTCACCTAGATCCT', 'TTCCGCGCACATTT', 'TTCCTGTGTGAAAT', 'TTCGATATCAAGCT', 'TTCGCGTTAAATTT', 'TTCGGAAAAAGAGT', 'TTCGGTCCTCCGAT', 'TTGCGCAACGTTGT', 'TTGCTGGCGTTTTT', 'TTGGAAAACGTTCT', 'TTGGCCGCAGTGTT','TTGGGAAGGGCGAT','TTGGTATCTGCGCT', 'TTGGTCATGAGATT', 'TTGGTCTGACAGTT', 'TTGTAAGCGTTAAT', 'TTGTTAAATCAGCT', 'TTGTTCCCTTTAGT', 'TTGTTGCCATTGCT', 'TTTAAATTAAAAAT', 'TTTATCAGGGTTAT', 'TTTCACCAGCGTTT', 'TTTCCCCGAAAAGT', 'TTTCGTTCATCCAT', 'TTTCTACGGGGTCT', 'TTTCTGTGACTGGT', 'TTTGGAACAAGAGT', 'TTTGGGGTCGAGGT', 'TTTGGTCATGAGAT', 'TTTTAAATCAATCT', 'TTTTCAATATTATT', 'TTTTTCAATATTAT', 'TTTTTCCATAGGCT'
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal EcoRI site found at 702
Illegal PstI site found at 696 - 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 702
Illegal PstI site found at 696 - 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 702
Illegal BamHI site found at 684 - 23INCOMPATIBLE WITH RFC[23]Illegal EcoRI site found at 702
Illegal PstI site found at 696 - 25INCOMPATIBLE WITH RFC[25]Illegal EcoRI site found at 702
Illegal PstI site found at 696
Illegal NgoMIV site found at 328 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 2107
Illegal SapI site found at 1024
| None |