Part:BBa_K1355001:Design
For this genetic construction, we followed these summarized steps in the following image:
Read more about the design of this genetic construction on the extended version below:
1) Transformation of DH5-alpha with the pBSK plasmid wich contains the BBa_K1355001 - bidirectional promotor regulated by the MerR protein.
2) Extraction and quantification of the BBa_K1355001 plasmid DNA;
3) Verifying the electrophoretic profile of the extracted plasmid DNA;
Figure 1: Electrophoretic profile of the BBa_K1355001 plasmid DNA.
4) Restriction enzyme digestion of the BBa_K1355001 in pBSK with EcoRI and PstI and of BBa_J04450 in pSB1C3 with EcoRI and PstI aiming to isolate the biobrick fragment and isolate the plasmid backbone, respectively.
5) Checking the electrophoretic profile of digested samples;
Figure 2: A) Electrophoretic profile of BBa_K1355001 digested with EcoRI + PstI; B) Electrophoretic profile of the BBa_J04450 digested with EcoRI + PstI.
6) Purification from agarose gel of the fragment (BBa_K1355001) and the pSB1C3 plasmid backbone (BBa_J04450);
7) Checking the electrophoretic profile of purified samples;
Figure 3: A) Electrophoretic profile of BBa_K1355001 (fragment) purified; B) Electrophoretic profile of BBa_J04450 (pSB1C3 plasmid backbone) purified.
8) Ligation of the pSB1C3 plasmid backbone digested with the fragment (BBa_K1355001) using T4 DNA ligase;
9) Transformation of the ligation in DH5-alpha;
10) Selection of 6 colonies of transformed DH5-alpha with ligation system grown in chloramphenicol;
11) Colony PCR of the 6 colonies using the VR - VF2 primers for pSB1C3 plasmid;
12) Checking the electrophorectic profile to obtain results showing that the amplified samples is the junction of BBa_K1355001 with pSB1C3 plasmid backbone;
Figure 4: (A) - (F) Colony PCR of 6 colonies transformed with ligation system.
13) Plasmid DNA extraction of the BBa_K1355001 in pSB1C3 amplified using VR - VF2 (A, B and C colonies);
14) Checking the electrophoretic profile;
Figure 5: Electrophoretic profile of the BBa_K1355001 in pSB1C3 extracted plasmid DNA.
15) Restriction enzyme digestion of BBa_K1355001 with EcoRI + PstI, only with EcoRI and only with PstI aiming to analyze the fragment size to be isolated (digestion with EcoRI + PstI) or the size of the linearized vector (only with EcoRI or PstI);
16) Checking the electrophoretic profile of the digested sample to obtain results showing that the isolated fragment (sample digested with EcoRI + PstI) is the BBa_K1355001 in pSB1C3 and that the linearized vector (sample digested only with EcoRI or PstI) is the junction of our biobrick in pSB1C3;
Figure 6: (A) Electrophoretic profile of the BBa_K1355001 do not digested; (B) digested only with EcoRI; (C) digested only with PstI; and (D) digested with EcoRI + PstI, respectively.
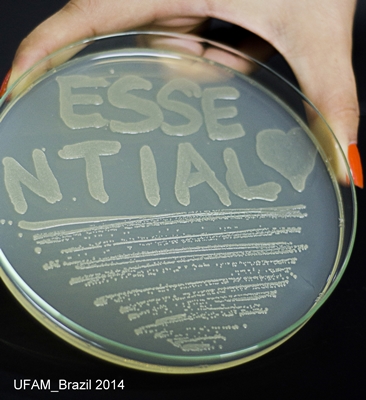
There is our new part - Essential Biobrick - Regulation and transport of Hg!
Figure 7: DH5-alpha transformed with Essential Biobrick (BBa_K1355001) in pSB1C3 grown in chloramphenicol;
The fragment - our biobrick BBa_K1355001 - in the digestion with EcoRI + PstI (sample D) has 1.234 base pairs and the vector pSB1C3 has 2.070 base pairs. The linearized vector contains about of 3.500 base pairs.
To finalize our molecular characterization - design, we also make the Sanger method of DNA sequencing.
Check it out our experience with this bidirectional promotor regulated by the MerR protein linked with the parts BBa_E0840, BBa_K1355000 and BBa_K346004 composing the parts device BBa_K1355002 (Hg bio-detector), BBa_K1355003 (Hg bio-accumulator) and BBa_K1355004 (Hg bioremediator)!