Part:BBa_K1123000
FNR Promoter
This part is a standard FNR promoter. It has two FNR binding sites at -41.5 and at -91.5 with 0 being the transcriptional starting point of this promoter. The promoter can be used in E.coli strains to induce protein expression in anaerobic environments. It is natively used to induce metabolic processes when the E.coli bacteria enter anaerobic environments. The part can be used to express any protein sequence placed behind it.
TU-Eindhoven Sequencing Results
This construct has been submitted to the iGEM registry and has been privately sequenced by the iGEM 2013 TU-Eindhoven. The sequencing results were positive indicating that the sequence and part submitted to the registry are correct. Sequencing was performed by BaseClear Netherlands
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 39
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterization
This part is one of the most essential parts used within the 2013 TU-Eindhoven iGEM project. Within our project we were focusing on the expression of MRI contrast providing proteins under anaerobic conditions. As we wished these anaerobic conditions to be the trigger for the protein expression we needed a promoter which would react to this environment and ultimately induce the protein expression. The FNR promoter is a naturally occurring promoter which exists in almost all strains of E.coli which makes it an extremely attractive system to use.
Upon entering a hypoxic environment the forming of [4Fe-4S]2+ dimers will be stimulated within FNR producing bacteria. These dimers are the transcription activators of the FNR promoter. They are able to bind to the promoter allowing the promoter to become active. Important to note is that the promoter we are using is a tandem promoter meaning that 2 [4Fe-4S]2+ dimers are able to bind to the promoter. This tandem effect enhances the induction effect of the promoter, ultimately leading to a higher expression rate.
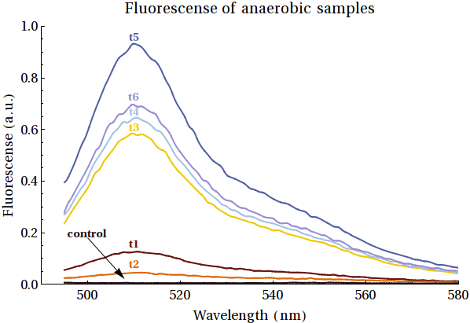
As it is not possible to quantify the workings of a promoter without expression we could only truly test this part in combination with a protein which we wished to express. To ease the analysis of the expression product we chose to express the EGFP protein over the CEST proteins we would eventually want to express. The expressions were performed first in BL21 bacteria however this did not provide us with the desired results. As we wished to show that our promoter did work, we attempted the same expression of EGFP only now in two other E.coli strains: XL-1-Blue and MG1655.
As it would turn out EGFP was formed in both these bacterial strains. Having said this it is our recommendation for other teams to attempt the expression in the XL-1-Blue bacteria over the MG1655 bacteria, as this gave higher yields that also increased over time. We should also mention that the yields of the expressed EGFP was rather low, even if we did see increased intensities over the course of time. The fluorescence measurements of the expression of EGFP samples over time can be seen below:
For a detailed analysis of the expressions of EGFP in both all three bacterial strains we refer you to the page of this composite part, where the FNR promoter has been combined with the EGFP protein, along with other functional DNA. We do this to allow you to see the entire construct we tested and not just the basic Promoter we used to achieve the expression.
Click Here to view the part BBa_K1123005 used for the expression.
NCKU_Tainan 2019 Characterization
Characterization
This year, we characterized FNR promoter (BBa_K1123000) which is an anaerobic promoter. Originally, we used this promoter to drive the expression of TAL constructs. However, finding that this promoter has a higher expression level of GFP under aerobic conditions, we did not use this promoter for our project. Instead, we used the strong constitutive promoter J23100 as our promoter.
We first cultured DH5α pSB1C3-Pfnr-GFP under aerobic and anaerobic conditions. After 10 hours of incubation, we fixed the E. coli cells in 2% agarose gel and placed it on a glass slide, then place the glass slide under a microscope for image capturing.
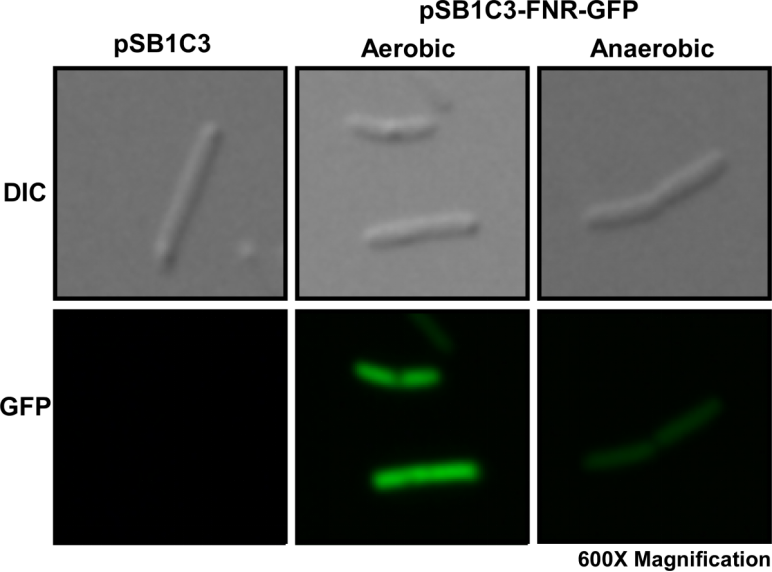
Fig. 1 Differential interference contrast (DIC) microscope image and fluorescence microscope image for a single E. coli cell.
We then compared the brightness of a single E. coli cell under a fluorescence microscope using ImageJ Intensity Processing. The results are shown below.

Fig. 2. Quantification of single E. coli cell (n=3) fluorescence intensity using ImageJ Intensity Processing after 10 hours of incubation in both aerobic and anaerobic conditions.
As seen in Fig. 2, after 10 hours of incubation, the fluorescence signal in single E. coli cell is significantly higher in aerobic conditions than in anaerobic conditions. Literature has reported that GFP requires oxygen molecules to fold properly[1] before it can emit fluorescence signal. We cannot exclude the possibility that in anaerobic culture, GFP protein is not folding properly and thus affecting the measurement result. However, we can still conclude this promoter is not anaerobic specific. It is recommended that future iGEMers should use other reporter genes that will not be affected by oxygen like luciferase to further characterize this Biobrick.
Refence
[1] Coralli, C., Maja Cemazar, Chryso Kanthou, Tozer, G. M., & Dachs, G. U. (2001). Limitations of the Reporter Green Fluorescent Protein under Simulated Tumor Conditions. Cancer Research, 61(12), 4784–4790.
| None |