Part:BBa_K1051502
Sic1 mutation, non stop codon
Purpose
Works for the cell synchronization device, to stop all yeast cells to G1 phase.
Principle
Sic1, a protein, is a stoichiometric inhibitor of Cdk1-Clb (B-type cyclins) complexes in the budding yeast Saccharomyces cerevisiae. Because B-type cyclin-Cdk1 complexes are the drivers of S-phase initiation, Sic1 prevents premature S-phase entry.Multisite phosphorylation of Sic1 is thought to time Sic1 ubiquitination and destruction, and by extension, the timing of S-phase entry.
Experimental results show that overexpression of Sic1 regulated by GAL1 promoter, the cells will stop at late G1, and the cell becomes larger than the original.
We try to mutate seven phosphorylation site of Sic1, it can help stopping the cell cycle in the G1 phase and further achieve synchronization.
<img src="http://upload.wikimedia.org/wikipedia/commons/0/06/Sic1_David_Morgan10-5.jpg" />
Engineering
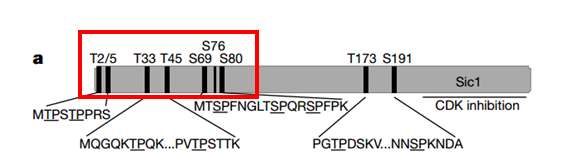
Seven N-terminal phosphorylation sites are mutated in our version to slow down the degradation. This is the version without stop codon.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 310
References
[1] T T Nugroho and M D Mendenhall.An inhibitor of yeast cyclin-dependent protein kinase plays an important role in ensuring the genomic integrity of daughter cells.Mol Cell Biol. 1994 May; 14(5): 3320–3328;
[2] Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M.Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication.Nature. 2001 Nov 29;414(6863):514-21;
[3] Audra Day, Colette Schneider, Brandt L. Schneider.Yeast Cell Synchronization.Methods in Molecular Biology™ Volume 241, 2004, pp 55-76.
| n/a | Sic1 mutation, non stop codon |
