Part:BBa_K1022102
pT7: RBS: His6- SUMO: Signiferin
This part codes for the peptide 'Signiferin' tagged with 'His-6-SUMO' molecule. Its production is triggered by IPTG (through pT7 promoter).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 46
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 170
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 355
- 1000COMPATIBLE WITH RFC[1000]
Characterization
For more info, visit [http://2013.igem.org/Team:TU-Delft/Peptides#SUMO Peptide production] on TU Delft iGEM13 Wiki
Peptide production
Introduction:
In the project by iGEM 13 Team TU Delft, a SUMO-peptide fusion was opted as a suitable expression system as they make the fusion proteins more soluble. The peptide by itself is not soluble in the cytoplasm but making a fusion of peptide with Small Ubiquitin like Modifiers (SUMO) will increase the solubility of the peptide, thus increasing the cytoplasmic fraction of the peptide.
A gene was constructed in such a way that the SUMO-peptide production was driven by the strong T7 phage promoter. This gene containing plasmid was harbored in a BL21(DE3) strain that has lac promoter driven T7 polymerase. Upon induction by IPTG the SUMO peptide fusion is produced as a soluble protein fraction.
The protocol can be seen [http://2013.igem.org/Team:TU-Delft/Protocol_10#protocol_10 here].
Result:
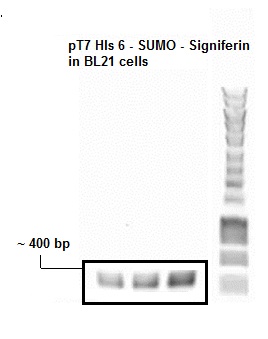
- Figure 1: Colony PCR SUMO Peptide
The presence of plasmid with gene inserts encoding the SUMO-peptide (BBa_K1022102) was confirmed by a colony pcr. The expected size of the insert with the T7 promoter was approximately around 400 bp which was clearly evident from the agarose gel picture. An Eurogentec Smartladder MW-1700-10 (https://secure.eurogentec.com/uploads/TDS-MW-1700-10.pdf) was used to identify the size of the fragments.
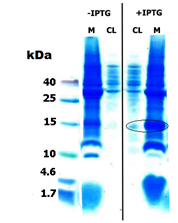
- Figure 2: Tricine gel of Uninduced and induced SUMO-Peptide CL: Cell lysate, M: Membrane pellet
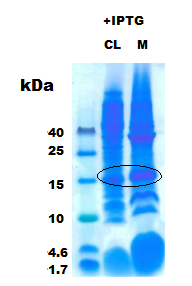
- Figure 3: SUMO-Peptide with IPTG induction CL: Cell lysate, M: Membrane pellet
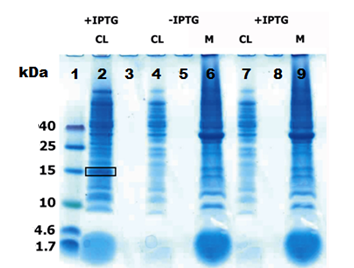
- Figure 4: SUMO-Peptide solubilty
Lane 1: Low molecular weight marker, Lane 2: IPTG induced SUMO-peptide cell lysate fraction (CL) Lane 4: Uninduced peptide-his without SUMO in cell lysate (CL), Lane 6:Uninduced peptide-his without SUMO in membrane pellet(M) Lane 7:Induced peptide-his without SUMO in cell lysate (CL), Lane 9:Induced peptide-his without SUMO in membrane pellet (M)
Then strains were subjected to protein expression by means of IPTG induction. The cell lysate and the centrifuged cell debris were run on a 16 % tricine gels at 90 V for 90 minutes. The figure 2 clearly shows a over-expression of our fusion protein at ~15 kDa both in whole cell lysate (CL) and the membrane (M), This was from just 10 µg of total protein for cell lysate (CL) and 30 µg of total protein in the membrane (M).
- Figure 5: Sequence coverage from Tandem MS [Red letters]
Then just the induced CL and M were run with 30 µg of total protein per well as in figure 3 the results were again consistent (Black circle). One of the over-expressed band was excised and subjected for in-gel tryptic digestion and then to MS/MS measurement in Q-trap MS instrument (Thermo Scientific). To prove the solubility of the peptide the the CL and M from constructs that express peptide without SUMO was run with the CL from SUMO- peptide as in figure 4.
Discussion:
The SUMO domain was added to the peptide to improve its solubility [1]. This was really evident from the figure (2, 3). The peptides without SUMOylation did not have any over-expression in the cell lysate which is clearly seen in the figure 4 [ lanes 4,6 (uninduced) 7,9 (induced)] . But when a SUMO fusion of the same peptide was expressed they appeared as a thick band in the cell lysate as in the figure 4 [lane 2,black box] . From the figures [2,3] it is evident that there was some fraction of protein left with the cell debris. This clearly explains that SUMO does not increase the solubility to 100 % but just enhances the fraction of the soluble protein, thus facilitating easy purification. The band which was subjected to the MS/MS measurements clearly gave a 95 % sequence coverage to SUMO-peptide (signiferin) as in Fig 5. Thus, we prove that the SUMO increases the solubility and also they are not toxic to the cell.
Highlights:
The SUMO domain that was used in our project was synthesized as a gene from DNA 2.0. The SUMO was designed to be bio-brick compatible without the restriction sites that are present in the prefix and suffix. The restriction sites where removed without disturbing the codons.
The Biobrick was designed in such a way that the ease of cloning SUMO fusion protein to be expressed has been improved. An Age I restriction site was introduced at the end of SUMO which when translated to amino acids Tyrosine Glycine Glycine. This is the recognition sequence of the Ulp-1 [2] protease that can cleave the peptides from the SUMO without any scar. So, one can use Age I and any one of the suffix enzyme to make a SUMO fusion.
Peptide Characterization (MIC)
Introduction:
An important part of the project is inhibition of growth or killing of bacteria with the use of antimicrobial peptides (AMPs). In order to get an idea of the toxicity of our peptides we conducted several minimal inhibiting concentration (MIC) experiments with synthetic peptides ordered from DNA 2.0. These MIC measurements where done on E. coli, B. subtilis and S. delphini, with the first as representative of our Gram-negative expression host, the second for the Gram-positive targets and the last for our specific target.
MIC determination:
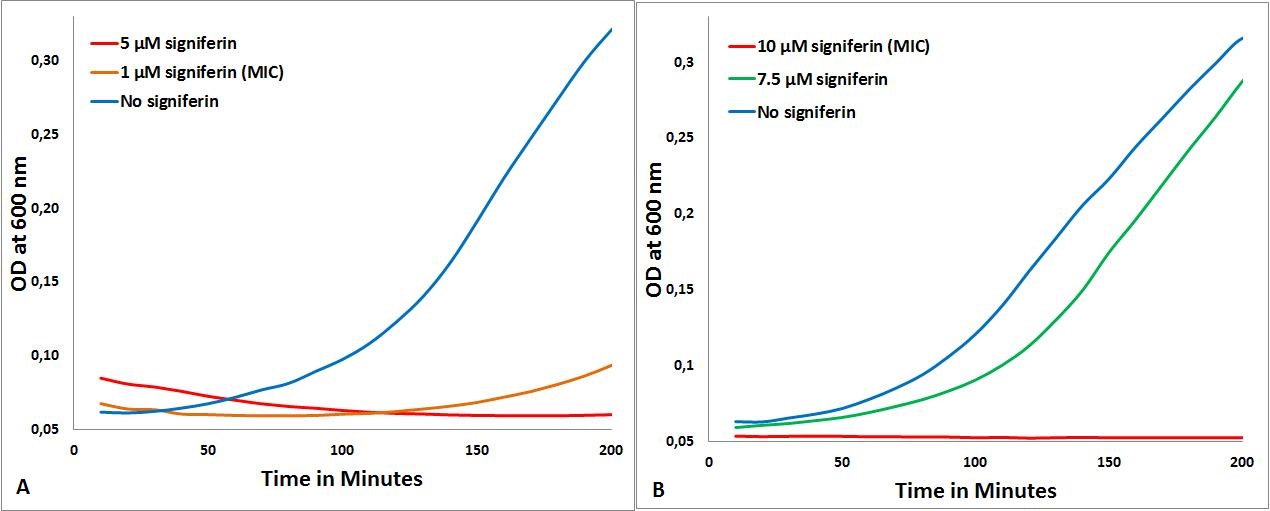
The MIC of signiferin on S. delphini, B. subtilis and E. coli were done according to the protocol described [http://2013.igem.org/Team:TU-Delft/PeptideCharacterization here]. S. delphini is the most sensitive for signiferin, as the MIC was determined to be 1µM (part A in figure below), no growth is seen at this concentration. The MIC for B. subtilis was determined to be 10µM (part B in figure below).
- MICs of signiferin. 1A: signiferin on S. delphini, 1B: signiferin on B. subtilis
The expected selectivity of the chosen peptides for Staphylococcus was confirmed by these experiments, as for all the peptides for which we could determine a MIC, that MIC was the lowest on S. delphini. For MIC by other peptides used in iGEM 2013 Team TU Delft, Click [http://2013.igem.org/Team:TU-Delft/PeptideCharacterization here].
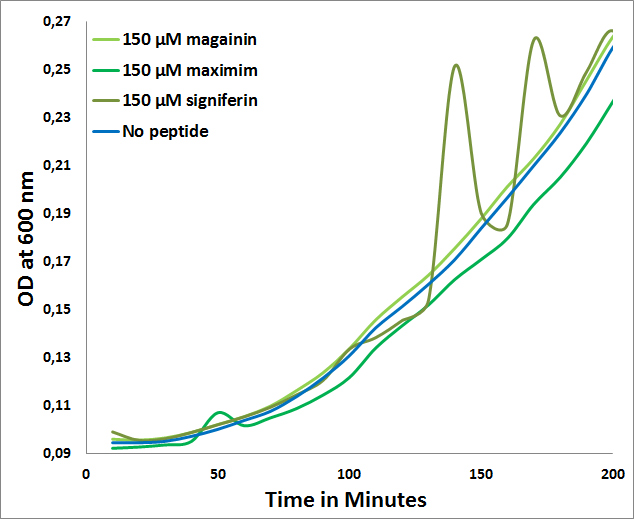
Comparison with two peptides:
The fact that for none of the peptides a MIC could be determined for E. coli (>150µM) further confirms the expected selectivity towards Gram-positives which is observed in the following graph:
- MICs of Maximim-H5, Signiferin and Magainin on E. coli
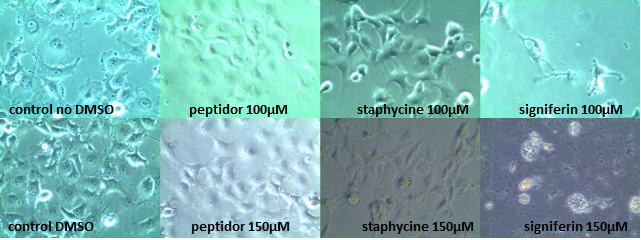
COS-1 toxicity determination:
In order to determine the toxicity of the AMPs on mammalian cells we tested the most promising peptides on COS-1 cells[3]. As signiferin, staphycine and peptidor gave the best results in the experiments of MIC, they were chosen to be tested with concentrations up to 150µM on COS-1 cells. COS-1 cells were chosen as they, because of their simian nature, strongly resemble human cells with respect to membrane properties and crucial cell functions and mechanisms[4]. Normally healthy COS-1 cells attach to the bottom of the well, showing a clear fibroblastic morphology[5]. When they start to die they will swell and eventually detach from the bottom forming spherical cells which upon lysis leave behind cell debris.
Signiferin showed to be toxic above 100µM, whereas staphycine and peptidor turned out not to be toxic up to concentrations of 150µM. Two controls are shown, as peptidor and signiferin are dissolved in water whereas staphycine is in 80% DMSO. All photos shown are taken 24 hours after induction, but are representative for the photos taken 4 hours after induction. In the 150µM signiferin experiment no live cells could be observed at both 4 hours and 24 hours after induction.
For more info, visit [http://2013.igem.org/Team:TU-Delft/PeptideCharacterization Peptide characterization] on TU Delft iGEM13 Wiki
SUMO-Peptide cleavage by Ulp1 protease
For more info, visit [http://2013.igem.org/Team:TU-Delft/Peptides#cleavage SUMO Cleaved by Ulp 1] on TU Delft iGEM13 Wiki
The construct used for testing this is: BBa_K1022117
References
1. Panavas T, Sanders C, Butt TR.. (2009). SUMO fusion technology for enhanced protein production in prokaryotic and eukaryotic expression systems. Methods in Molecular Biology. 10 (8), p4-20.
2. Lee, C., Sun, H., Hu, S., Chiu, C., Homhuan, A., Liang, S., Leng, C. and Wang, T. 2008. An improved SUMO fusion protein system for effective production of native proteins. Protein Science, 17 (7), pp. 1241--1248.
3. J.F. Hancock, “COS Cell Expression”, Methods in Molecular Biology. Vol 8 pp 153-158, 1992.
4. Fred C. Jensen, Anthony J. Girardi, et al., “Infection of Human and Simian Tissue Cultures with Rous Sarcoma Virus”, Proc Natl Acad Sci U S A. 1964 July; 52(1): 53–59. Jul 1964.
5. P. Shashidharan, G.W. Huntley, et al., “Immunohistochemical localization of the neuron-specific glutamate transporter EAAC1 (EAAT3) in rat brain and spinal cord revealed by a novel monoclonal antibody, Brain Research. Volume 773, Issues 1–2, Pages 139–148, Okt 1997.
| n/a | pT7: RBS: His6- SUMO: Signiferin |