Difference between revisions of "Part:pSB1A3"
Smelissali (Talk | contribs) (→Usage and Biology) |
Smelissali (Talk | contribs) |
||
| Line 17: | Line 17: | ||
*Also, as a general comment, the dsDNA that is listed when sequence information is requested is impossible to work with in most DNA analyzer programs (e.g. NEBcutter) since these programs expect top strand only. It would be IDEAL if the dsDNA could be listed as ss on demand. | *Also, as a general comment, the dsDNA that is listed when sequence information is requested is impossible to work with in most DNA analyzer programs (e.g. NEBcutter) since these programs expect top strand only. It would be IDEAL if the dsDNA could be listed as ss on demand. | ||
**If you click the "get sequence as shown" link in the "sequence and features" section of a part's page, you can get a ssDNA text file (smile)! [Drew] | **If you click the "get sequence as shown" link in the "sequence and features" section of a part's page, you can get a ssDNA text file (smile)! [Drew] | ||
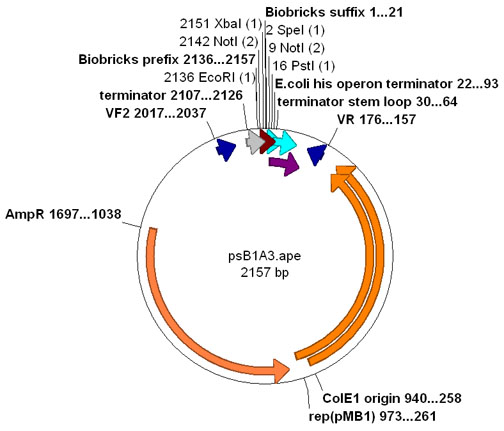
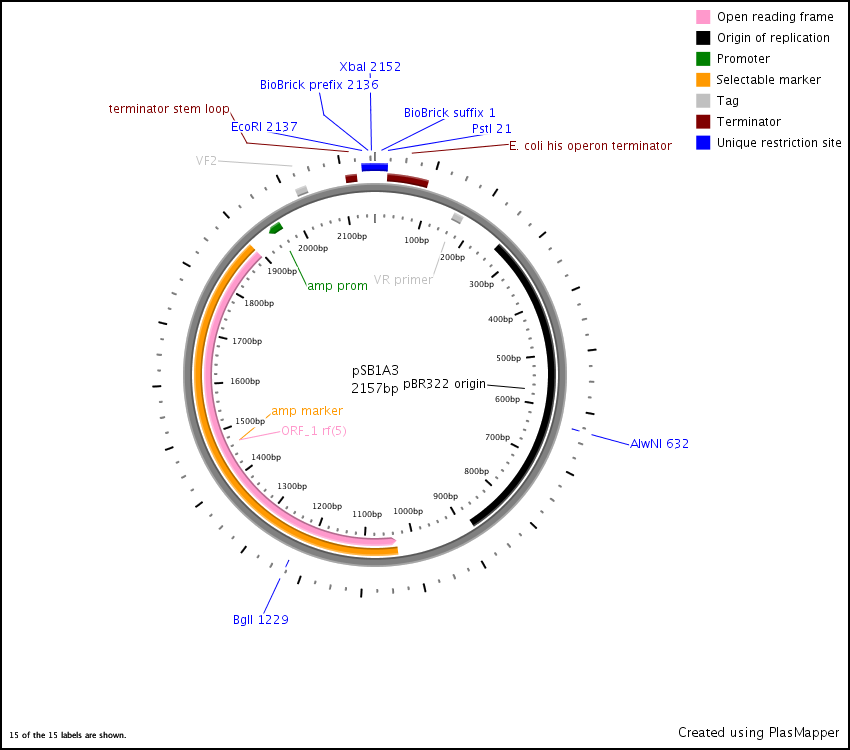
| − | **Annotated [http://www.biology.utah.edu/jorgensen/wayned/ape/ APE] file for [https://static.igem.org/mediawiki/parts/e/e4/PsB1A3.ogg pSB1A3] [[Image:Psb1A3 map.jpg|thumb|center|pSB1A3 Plasmid Map]] | + | **Annotated [http://www.biology.utah.edu/jorgensen/wayned/ape/ APE] file for [https://static.igem.org/mediawiki/parts/e/e4/PsB1A3.ogg pSB1A3] [[Image:Psb1A3 map.jpg|thumb|center|pSB1A3 Plasmid Map]][[Image:PlasMap pSB1A3 plasmapper.png|thumb|center|psB1A3 Plasmapper]] |
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
Latest revision as of 17:22, 11 June 2007
High copy BioBrick assembly plasmid
pSB1A3-1 is a high copy number plasmid carrying ampicillin resistance.
The replication origin is a pUC19-derived pMB1 (copy number of 100-300 per cell).
pSB1A3 has terminators bracketing its MCS which are designed to prevent transcription from *inside* the MCS from reading out into the vector. The efficiency of these terminators is known to be < 100%. Ideally we would construct a future set of terminators for bracketing a MCS that were 100% efficient in terminating both into and out of the MCS region.
Usage and Biology
- This plasmid is not giving the predicted EcoRI/PstI digest. From the description at the registry site, there should be an EcoR1 at bp119 and a PstI at 156. I see a band that is btw 500 and 600 bps.
- Primers for these Biobrick vectors can be found in Part:BBa_G00100 (aka VF2) and Part:BBa_G00101 (aka. VR)
- Also, as a general comment, the dsDNA that is listed when sequence information is requested is impossible to work with in most DNA analyzer programs (e.g. NEBcutter) since these programs expect top strand only. It would be IDEAL if the dsDNA could be listed as ss on demand.
- If you click the "get sequence as shown" link in the "sequence and features" section of a part's page, you can get a ssDNA text file (smile)! [Drew]
- Annotated [http://www.biology.utah.edu/jorgensen/wayned/ape/ APE] file for pSB1A3
Sequence and Features
Assembly Compatibility:
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 2134
Illegal SpeI site found at 2
Illegal PstI site found at 16
Illegal NotI site found at 9
Illegal NotI site found at 2140 - 21INCOMPATIBLE WITH RFC[21]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 2134 - 23INCOMPATIBLE WITH RFC[23]Illegal prefix found at 2134
Illegal suffix found at 2 - 25INCOMPATIBLE WITH RFC[25]Illegal prefix found at 2134
Plasmid lacks a suffix.
Illegal XbaI site found at 2149
Illegal SpeI site found at 2
Illegal PstI site found at 16 - 1000INCOMPATIBLE WITH RFC[1000]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal BsaI.rc site found at 1173