Part:BBa_K883001
CCMV wt coat protein under IPTG induced promoter
[http://2012.igem.org/Team:Wageningen_UR/ModifyingtheCCMV CCMV] is an abbreviation of the Cowpea Chlorotic Mottle Virus, one of the most studied viruses in producing [http://2012.igem.org/Team:Wageningen_UR/VLPs Virus Like Particles] (VLPs). This part is the coding sequence of the coat protein of the CCMV virus. When expressed in Escherichia coli, these coat proteins can be harvested and be assembled in vitro using the Wageningen_UR iGEM 2012 protocols. This part is provided for the iGEM community so that they can use this part to produce these VLPs for their one own use, including but not limited to packaging, making fusion proteins to the N/C terminal. This all for to be used in a wide variety of applications. for ideas what kind of applications can be mediated with CCMV VLPs, check http://2012.igem.org/Team:Wageningen_UR/Applications
Background
This part is made from the wild type CCMV coat protein. CCMV is the abbreviation of Cowpea Chlorotic Mottle Virus, and is a well studied black eyed pea infecting virus. It is a virus in the family Bromoviridae, and it is one of the most studied plant viruses. As from the discovery of the virus in 1967, it was an interesting virus due to the easy cultivation and harvesting procedures involved. Interest in this virus is especially in the ability to disassemble the virus, remove the content and refold the empty particle simply by altering the pH. This makes this virus one of the first and best studied Virus Like Particles.
Usage and Biology
Protocols for production and purification:
- [http://2012.igem.org/Team:Wageningen_UR/Protocol/StartupCCMV CCMV production protocol part 1]
- [http://2012.igem.org/Team:Wageningen_UR/Protocol/DialysisCCMV CCMV production protocol part 2]
- [http://2012.igem.org/Team:Wageningen_UR/Protocol/RoundupCCMV CCMV purification protocol]
Results
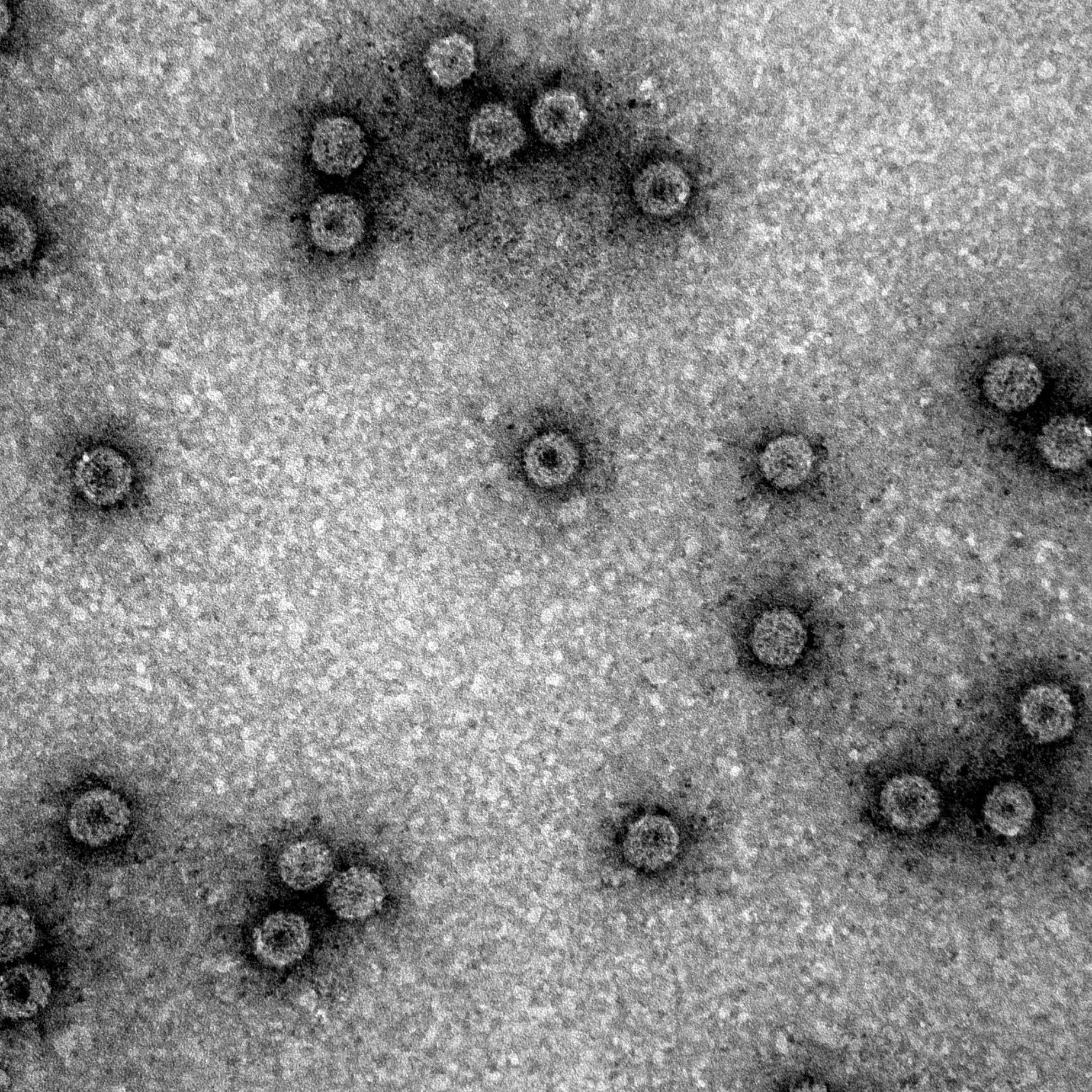
In order for us to be a good and functional part, we want to know if VLPs are formed with this brick using the protocol from above. We used two methods to detect the VLPs, Electron Microscopy (EM) and Dynamic Light Scattering (DLS). These methods are described on the [http://2012.igem.org/Team:Wageningen_UR/MethodsDetection detection page] of the Wageningen UR 2012 iGEM team.
In figure 1 it shows that with the protocol it is possible to form VLPs of the wild-type CCMV monomers. This means we have a viable part and a working protocol to produce VLPs.
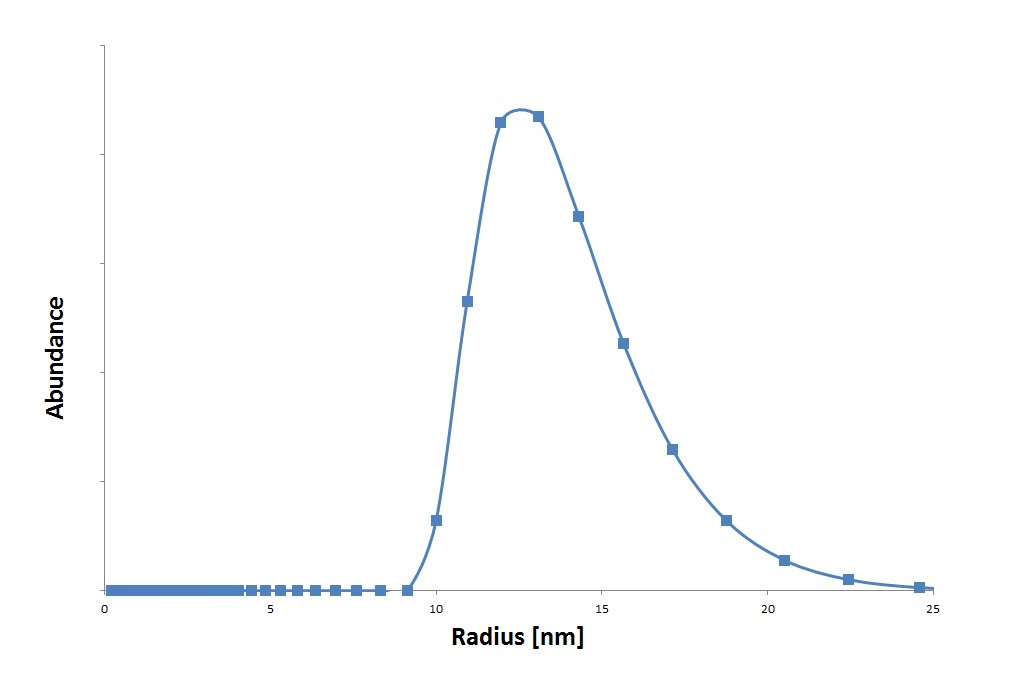
Next to Electron Microscopy we used Dynamic Light Scattering, With this technique we were able to get an another conformation of VLP formation, strenghtening that this part is working and that the protocol is able to produce VLPs. Besides detection with the DLS we also made a size distribution graph, seen in graph 1, from the raw data.
It shows that the predicted range is around 15 nm, which is roughly the size of the radius of the wild-type CCMV.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 368
- 1000COMPATIBLE WITH RFC[1000]
Functional Parameters
CCMV is a virus that can be useful in nanotechnology, this is because it is a well-studied virus and certain physical properties of the CCMV-VLP are already known. We want to provide additional information about the conditions in which the CCMVs are stable. These conditions are important when it comes to site-specific drug delivery. A better understanding of what will happen when the VLPs are injected into the bloodstream, will result into a better treatment for patients.
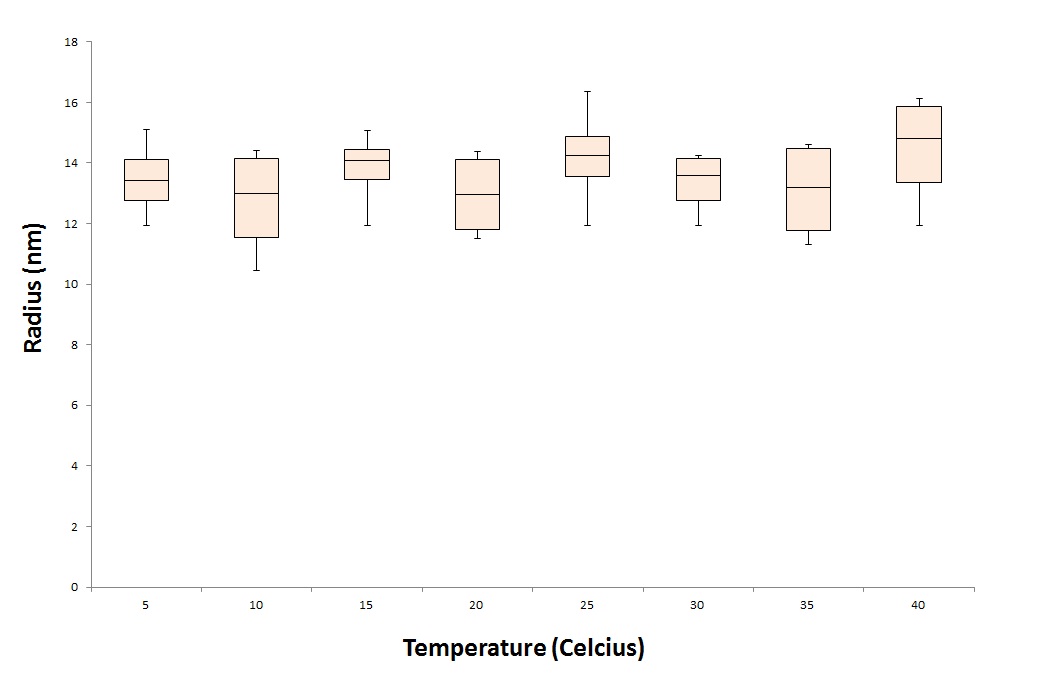
To test the stability of the CCMV VLPs, we treat the VLPs under different conditions. One parameter we want to test is, the temperature stability of the CCMV VLPs. To test this we put the CCMV VLPs for 24 hours in waterbaths with different temperatures. These temperatures range from 5 degrees celcius to 40 degrees representing the ill human being. After the treatment we tested the samples under the DLS and EM.
As seen in graph 2 and figure 2, the particles are still detected after a 24 hour treatment with 40 degrees Celcius. This means that the particles will not break down immidatly when exposed to the temperature of an ill human being.
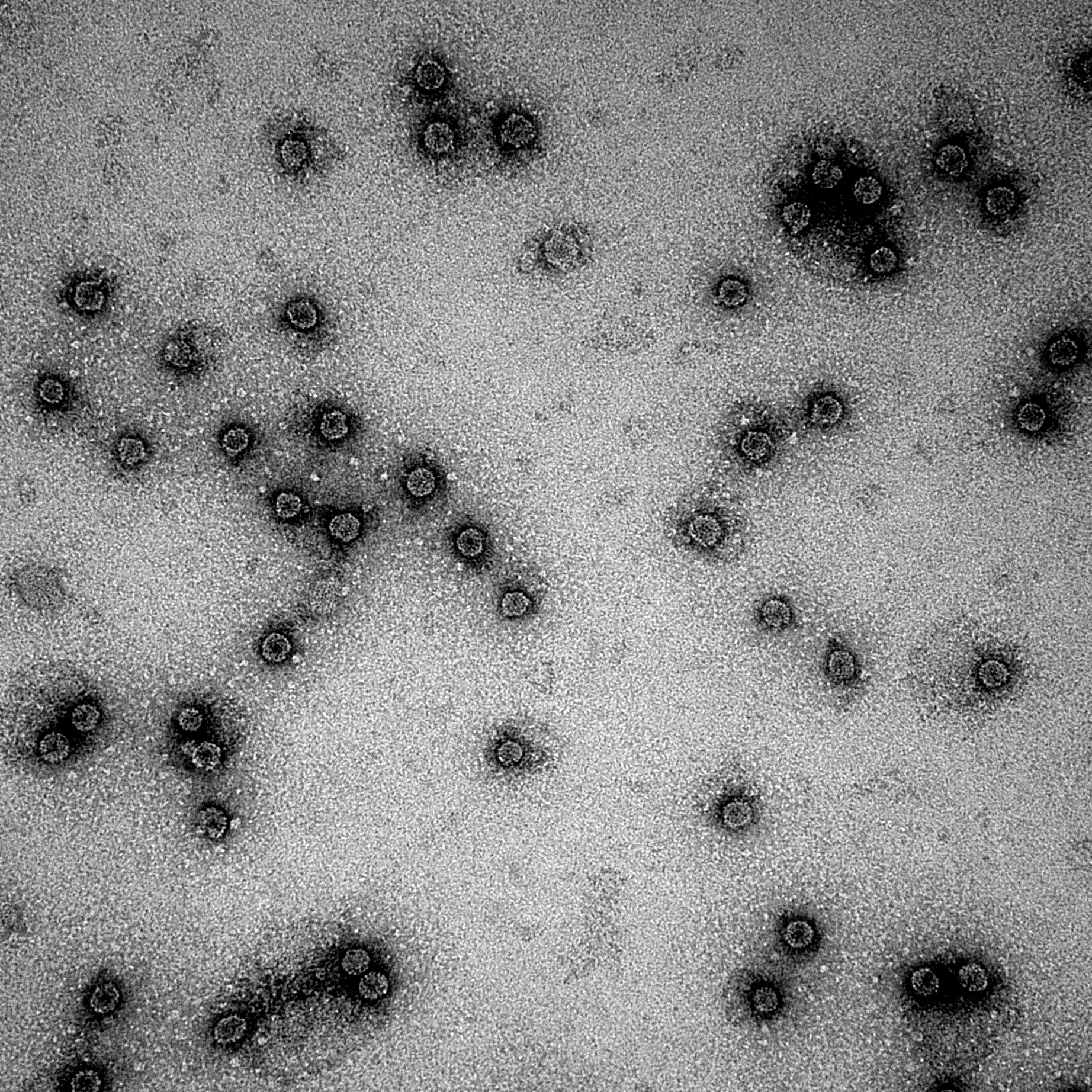
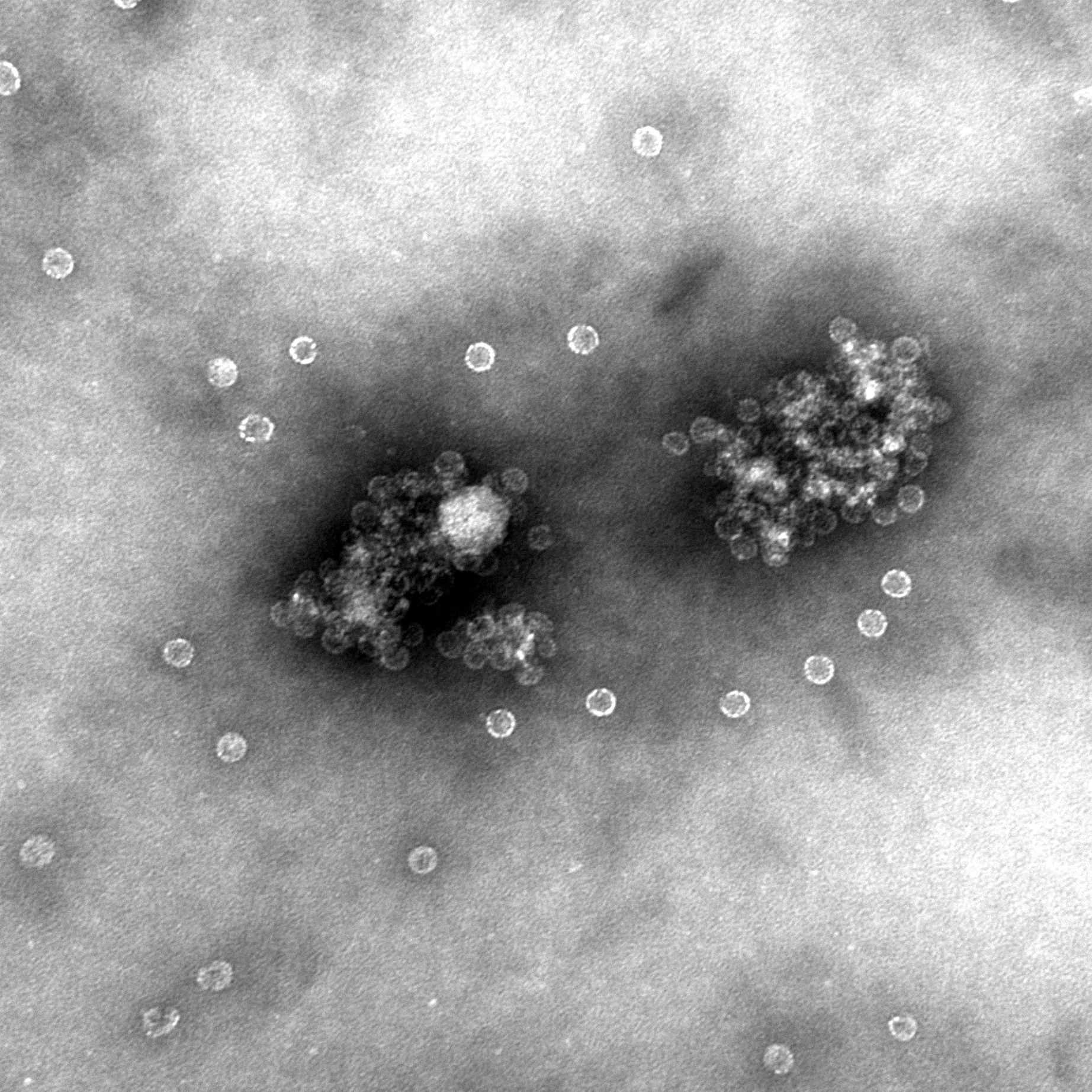
CCMV VLPs are renown for its wide pH stability. It is stable between a pH range of 4.7 and 6.5. At a pH of 4.7 the CCMV is in it's unswollen form with a radius of around 14 nm. At a pH of 6.5 it is in it's swollen form, the VLPs radius is around 15-16nm. It is possible to change form, but change the pH gradually with the use of dialysis. If you change the pH too sudden, the VLPs are falling apart and forms aggregates as seen in figure 3.
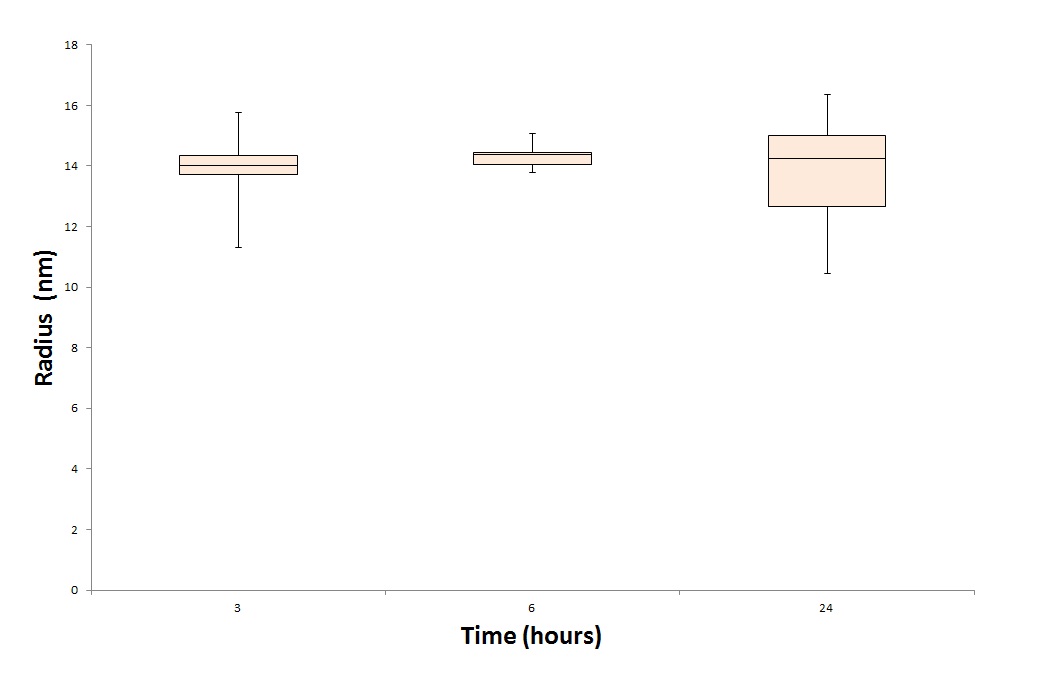
To test the stability of the VLPs during a long period of time, we deviced a experiment. We produced multiple CCMV VLPs and measure them after 3, 6 and 24 hours after formation.
In theory, under perfect conditions (pH at 4.7, T = 4 degrees Celcius) it is possbile to store the CCMV VLPs for a good few weeks. Be aware that aggregate formation may occure when the VLPs are exposed to changing pH or temperature.
*Safety notice*
This part is isolated from a virus and, when assembled, will form particles that very much resemble this virus in size and shape. However, no additional viral information is stored in this part and viral infection and/or replication can therefore be ruled out. It is completely safe for use in normal laboratory circumstances.
| None |