Part:BBa_K864601
|
Lambda t1 transcriptional terminator
|
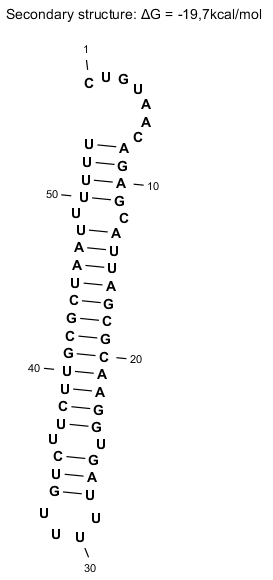
Secondary Structure
Contribution by NYU Abu Dhabi iGEM Team
Introduction:
The terminator t1 is located approx. 280 nucleotides beyond the int gene of bacteriophage k with role of both transcription terminator as well as provider of stability achieved by protecting int message from 3' exonucleolytic degradation [1]. Studies investigating t1 sequence in transcription termination and RNA stability induced point mutations in order to map G + C-rich region of dyad symmetry in the terminator and decrease its transcriptional termination [1]. Additionally, the tI mutations cause upstream transcript instability in vivo which is compensated by the host mutant deficient in polynucleotide phosphorylase resulting in increased steady state levels of these mutant transcripts [1]. These results show that the intact hairpin of tI is essential for efficient transcription termination and for maintaining mRNA stability by blocking the 3' to 5' exonucleolytic activity of polynucleotide phosphorylase [1].
Characteristics:
Lambda tI is an intrinsic terminator, with an interrupted dyad symmetry followed by a run of uridine residues. Percentage thymine of tI is increased when the MgCl2 concentration is decreased in the in vitro reaction which is consistent with other intrinsic terminators [2]. However, λ tI has an unusual structure in that the hairpin helix is almost 50% A-T and G-C base pairs, and the loop contains a sequence of 5 U residues [4]. Despite that they are multiple, only one to two termination sites are seen upon their mapping. This is unusual since part of the stem is mainly A-U and therefore the secondary structure is mainly found within the RNA hairpin stem [2]. Lastly, tI termination sites occur in the uridine residues with some heterogeneity, and they overlap with a retroregulation structure that triggers RNA processing by RNase III and PNPase [2]. Therefore, tI is a complex transcriptional terminator where protein-terminator interactions might be expected [2].
Discovery:
The tI point mutations reduced transcription termination at tI and induced transcript instability of those messenger RNAs that do terminate at tI [1]. The in vivo stability of the tI mRNA is determined by the intact secondary structure of tI and the presence of an active PNPase [1]. Stem Loop structure of tI is necessary for both transcription termination and RNA stability, and point out the important role of tI int expression [1]. The 3’-untranslated trailer region af the int mRNA is unusually long [3].
References:
- Cisneros, B., Court, D., Sanchez, A., & Montafiez, C. (1996). Point mutations in a transcription terminator, λtI, that affect both transcription termination and RNA stability. Gene, 181(1-2), 127-133.
- Bermúdez-Cruz, R. M., Chamberlin, M. J., & Montañez, C. (1999). Nus A is involved in transcriptional termination on λ tI. Biochimie, 81(7), 757-764.
- Schmeissner, U., McKenney, K., Rosenberg, M., & Court, D. (1984). Transcription terminator involved in the expression of the int gene of phage lambda. Gene, 28(3), 343-350.
//direction/forward
| biology | |
| direction | Forward |
| forward_efficiency | 0.97 |