Difference between revisions of "Part:BBa K823049"
| Line 2: | Line 2: | ||
<partinfo>BBa_K823049 short</partinfo> | <partinfo>BBa_K823049 short</partinfo> | ||
| − | This composite part expresses a | + | This composite part expresses a GFP fusion protein on <i>B. subtilis</i> spores. |
| + | |||
| + | The LMU-Munich 2012 team aimed to display proteins on spores for usage as ''beads.'' | ||
| + | |||
| + | <p align="justify">For creating fusion proteins on <i>Bacillus</i> spores, the crust protein [[Part:BBa_K823031|''cotZ -2aa]] were brought into Freiburg Standard for in-frame translation and put under the control of the native spore crust promoter [[Part:BBa_K823030|P<sub><i>cotYZ</i></sub>]]. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <br> | ||
| + | |||
| + | |||
| + | |||
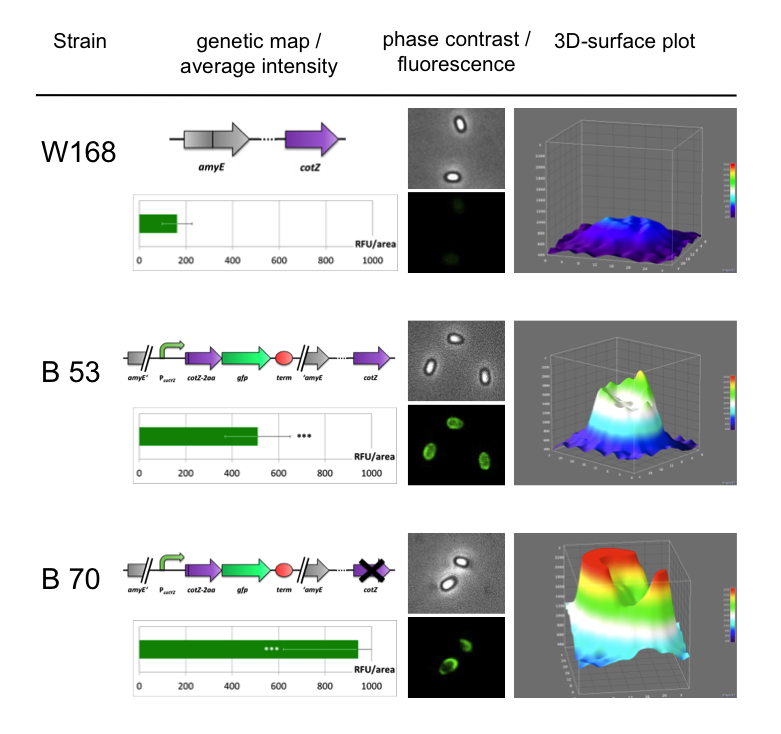
| + | All the '''Sporo'''beads were investigated by fluorescence microscopy and analysed with ImageJ and the statistical software '''R'''. The intensity bar charts show the fluorescence intensity, while the 3D graphs illustrate the fluorescence intensity spread across the spore surface, which correlates with the distribution of our fusion proteins. For analysis we measured the fluorescence intensity of a area of 750px per spore by using ImageJ and evaluated it with the statistical software R. The following graph shows the results of microscopy and ImageJ analysis. | ||
| + | </p> | ||
| + | |||
| + | [[File:LMU gfp spore data.png|620px]] | ||
| + | |||
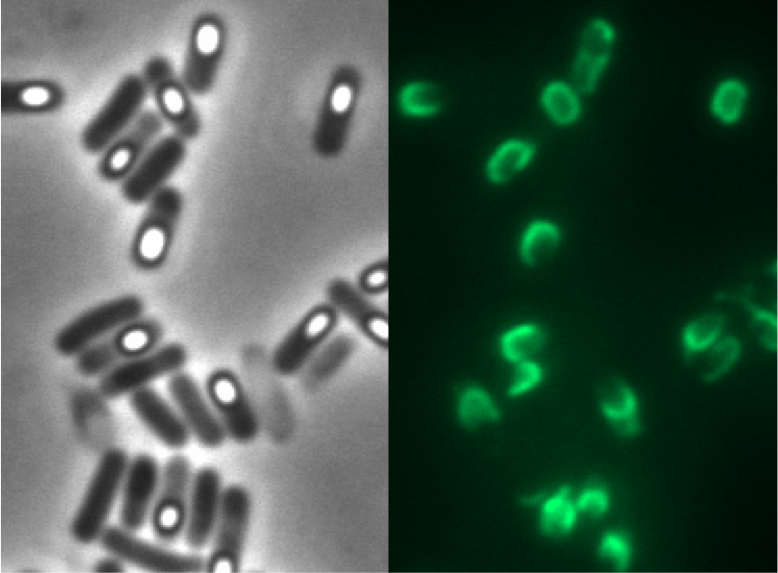
| + | <p align="justify">It appears the strain B70 with the construct [https://parts.igem.org/Part:BBa_K823049 P<sub>''cotYZ''</sub>-''cotZ''<sub>-2aa</sub>-''gfp''-''terminator''] and the deleted native ''cotZ'' had the strongest fluorescence. Thus this would be the strain for future '''Sporo'''beads with special functions. In some 3D graphs the picked cell does not respresent the average fluorescence intensity depicted in the bar chart, because it was chosen randomly.</p> | ||
| + | |||
[[Image:Glowing spore.png|thumb|center|500px|brightfield/fluerescence. Composite part was integrated in B. subtilis W168]] | [[Image:Glowing spore.png|thumb|center|500px|brightfield/fluerescence. Composite part was integrated in B. subtilis W168]] | ||
| + | whereas we created two different versions of the crust proteins. The restriction site NgoMIV was inserted just after the startcodon of the gene of the crust protein. Since this restriction site adds six additional basepairs the resulting gene is two codons longer [https://parts.igem.org/wiki/index.php?title=Part:BBa_K823032 CotZ]. It is not know if this insertion has any effect on protein expression that is why we created an additional version in which we deleted the following six basepairs, [https://parts.igem.org/wiki/index.php?title=Part:BBa_K823031 CotZ-2aa]. | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Revision as of 16:44, 26 September 2012
PcotYZ - cotZ (-2aa) - gfp - Terminator
This composite part expresses a GFP fusion protein on B. subtilis spores.
The LMU-Munich 2012 team aimed to display proteins on spores for usage as beads.
For creating fusion proteins on Bacillus spores, the crust protein cotZ -2aa were brought into Freiburg Standard for in-frame translation and put under the control of the native spore crust promoter PcotYZ.
All the Sporobeads were investigated by fluorescence microscopy and analysed with ImageJ and the statistical software R. The intensity bar charts show the fluorescence intensity, while the 3D graphs illustrate the fluorescence intensity spread across the spore surface, which correlates with the distribution of our fusion proteins. For analysis we measured the fluorescence intensity of a area of 750px per spore by using ImageJ and evaluated it with the statistical software R. The following graph shows the results of microscopy and ImageJ analysis.
It appears the strain B70 with the construct PcotYZ-cotZ-2aa-gfp-terminator and the deleted native cotZ had the strongest fluorescence. Thus this would be the strain for future Sporobeads with special functions. In some 3D graphs the picked cell does not respresent the average fluorescence intensity depicted in the bar chart, because it was chosen randomly.
whereas we created two different versions of the crust proteins. The restriction site NgoMIV was inserted just after the startcodon of the gene of the crust protein. Since this restriction site adds six additional basepairs the resulting gene is two codons longer CotZ. It is not know if this insertion has any effect on protein expression that is why we created an additional version in which we deleted the following six basepairs, CotZ-2aa.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 237
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 835