Part:BBa_K823031
cotZ: B. subtilis spore crust protein (-2aa)
This part codes for one of the outer most spore crust proteins in B. subtilis. We designed this part in the Freiburg standard (Assembly 25) to use it as a platform for the display of fusion proteins on B. subtilis spores.
Usage and Biology
The aim of the LMU-Munich 2012 project is to create spores that display fusion proteins on their surface. There are several different proteins forming the spore coat layers of B. subtilis spores. The outermost layer, the so called spore crust, is composed of CotZ and CgeA ([http://www.ncbi.nlm.nih.gov/pubmed?term=imamura%20et%20al.%202011%20spore%20crust Imamura et al., 2011]). Therefore, we used them to create functional fusion proteins to be expressed on our Sporobeads.
|
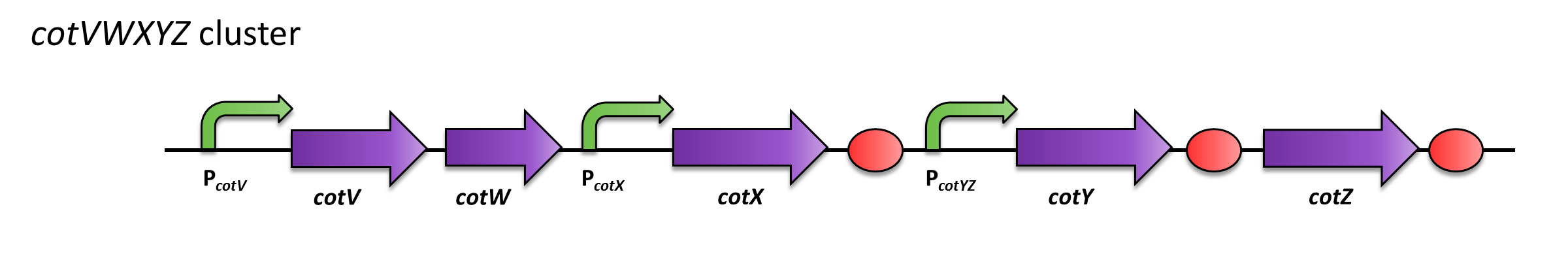
The cluster cotVWXYZ contains the gene cotZ which is cotranscribed with cotY. Expression is regulated by the promoter PcotYZ.
|
Evaluation
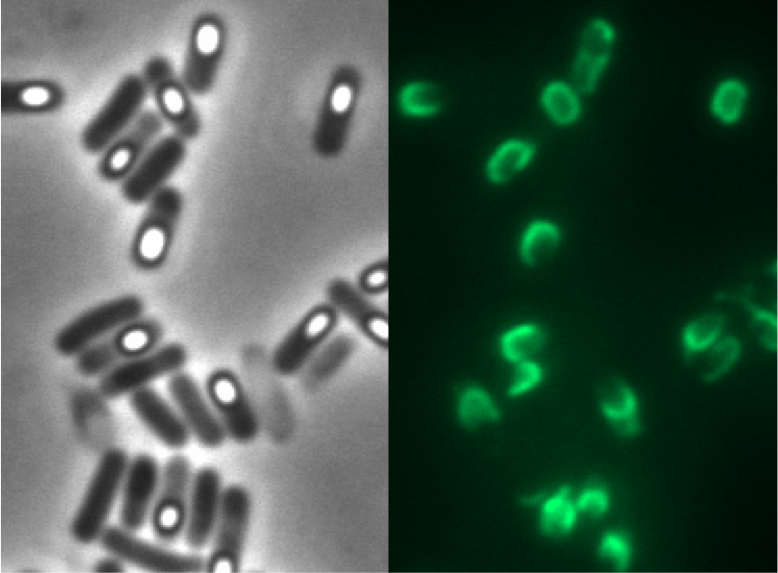
For our project the first step was to fuse gfp to cgeA and cotZ as a proof of principle. Thereby, we could determine if it is possible to display proteins on the spore crust and if their expression has any effect on spore formation.Finally, we were able to display GFP on B. subtilis spores by fusing it to CotZ (see device BBa_K823049).
NOTE: There are two versions of the cotZ gene in the registry, cotZ -2aa (this part), and cotZ. For our purpose, cotZ -2aa worked better.
For creating fusion proteins for the Sporobeads, the genes gfp, cotZ and cgeA were brought into Freiburg Standard whereas we created two different versions of the crust proteins. The restriction site NgoMIV was inserted just after the startcodon of the gene of the crust protein. Since this restriction site adds six additional basepairs, the resulting gene is two codons longer than the native cotZ. It was not know if this insertion has any effect on protein expression. Therefore we created two versions, this one, where we deleted two six basepairs, cotZ -2aa, and another, "cotZ" that is 2 aa longer.
See all the details about the LMU-Munich 2012 project [http://2012.igem.org/Team:LMU-Munich/Spore_Coat_Proteins here].
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 42
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| None |

![Composition of the Bacillus subtilis spore coat ([http://www.ncbi.nlm.nih.gov/pubmed?term=mckenney%202010%20spore%20crust McKenney et al., 2010] & [http://www.ncbi.nlm.nih.gov/pubmed?term=imamura%20et%20al.%202011%20spore%20crust Imamura et al., 2011])](/wiki/images/6/64/LMU_spore_crust.png)