Difference between revisions of "Part:BBa K823001"
(→Evaluation) |
|||
| Line 13: | Line 13: | ||
===Evaluation=== | ===Evaluation=== | ||
| − | This part was also evaluated in the publication [http://www.jbioleng.org/content/7/1/29 The Bacillus BioBrick Box: generation and evaluation of essential genetic building blocks for standardized work with Bacillus subtilis] by Radeck ''et al.''. | + | This part was also evaluated in the publication [http://www.jbioleng.org/content/7/1/29 The ''Bacillus'' BioBrick Box: generation and evaluation of essential genetic building blocks for standardized work with ''Bacillus subtilis''] by Radeck ''et al.''. |
===Luminescence measurements=== | ===Luminescence measurements=== | ||
Revision as of 16:54, 3 February 2014
PliaI promoter of Bacillus subtilis
PliaI is the promoter of the liaHI operon of B. subtilis. It has a very low basal activity and is inducible by bacitracin in a concentration dependant manner with a dynamic range over 100-fold.
This fragment does not contain a ribosome binding site.
Usage and Biology
PliaI is an inducible promoter from B. subtilis which is strongly induced by sublethal amounts of antibiotics that interfere with integrity and biosynthesis of the cell wall [http://www.ncbi.nlm.nih.gov/pubmed/15273097 Mascher et. al., 2004]). In the presence of a suitable stimulus, the two-component system LiaRS is activated. The activated response regulator LiaR binds to the operator of the promoter and induces the transcription of the lia locus. When the promoter is turned on the two proteins encoded by the liaHI operon are expressed [http://www.ncbi.nlm.nih.gov/pubmed?term=Journal%20of%20Bacteriology%2C%20188%20%2814%29%3A%205153%E2%80%935166: (Jordan et al., 2006)]. This promoter is evaluated with the reporters lux as well as lacZ. For more details visit the [http://2012.igem.org/Team:LMU-Munich/Data/Inducible Data] page of the LMU-Munich Team 2012 or get an overview of our whole project [http://2012.igem.org/Team:LMU-Munich Beadzillus].
Evaluation
This part was also evaluated in the publication [http://www.jbioleng.org/content/7/1/29 The Bacillus BioBrick Box: generation and evaluation of essential genetic building blocks for standardized work with Bacillus subtilis] by Radeck et al..
Luminescence measurements
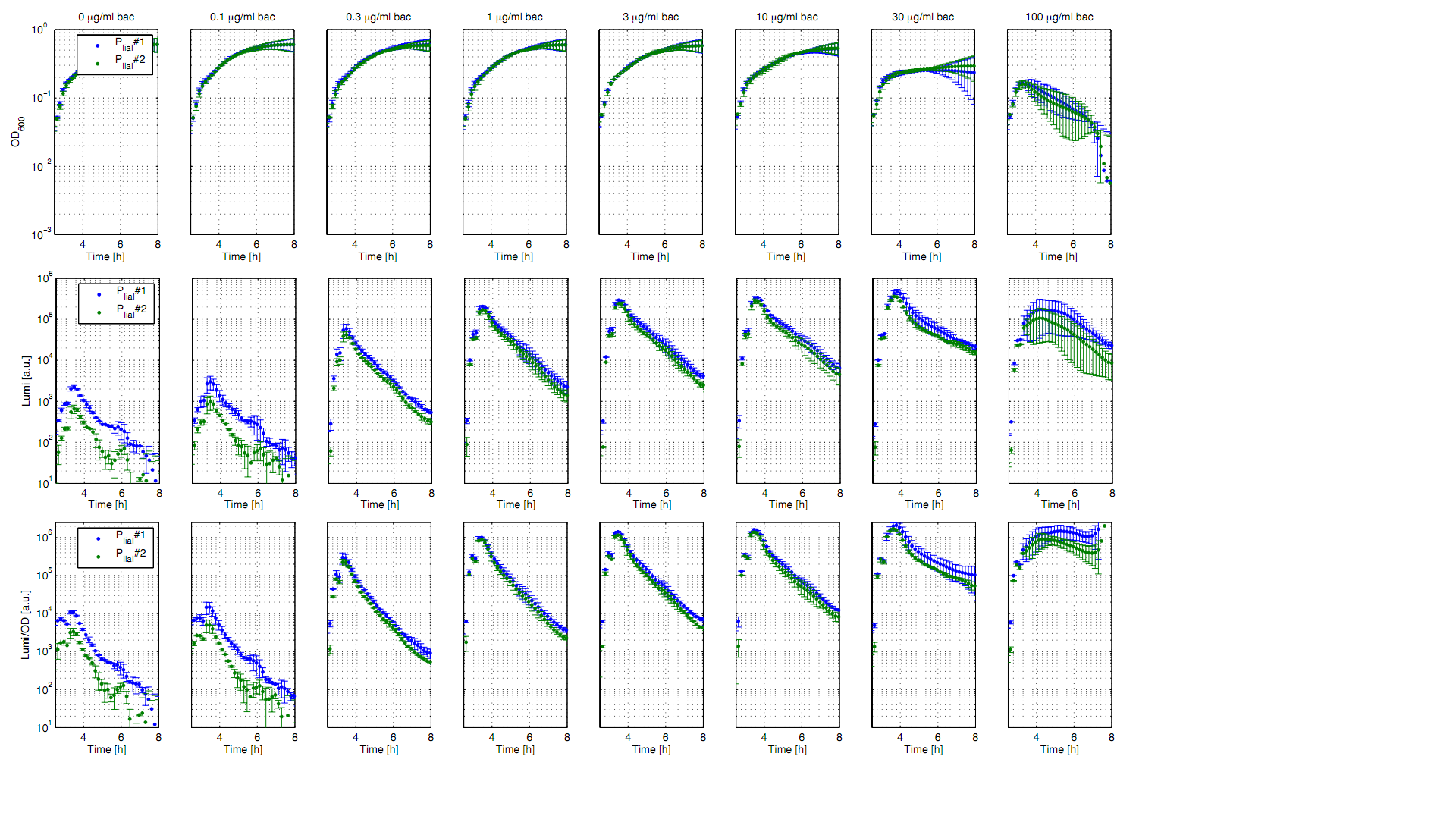
The bacitracin-inducible promoter PliaI was evaluated in the reporter vector pSBBs3C-luxABCDE which contains the lux operon. The promoter activity leads to gene expression and to the production of the protein luciferase. The luminescence produced by this protein can be measured with suitable microtiter plate reader as shown in Fig.1.
|
All clones show a normal growth behaviour up to a bacitracin concentration of 10 μg/ml. At higher concentrations of bacitracin the growth curves decrease because the cells lyse. The promoter PliaI shows a basal activity of about 10.000 Lumi/OD600. After induction with bacitracin the Lumi/OD600 increases depending of the concentration of bacitracin. The highest activity of about 1.5 Mio Lumi/OD600 can be measured after induction with 10-30 μg/ml bacitracin. To see the induction of this inducible promoter in comparison to an uninducible promoter we also did induction experiments with PliaG, which should not be inducible by bacitracin. As expected there is no response to the induction with bacitracin and PliaG shows a constant maximum of about 10.000 Lumi/OD600 (Data not shown).
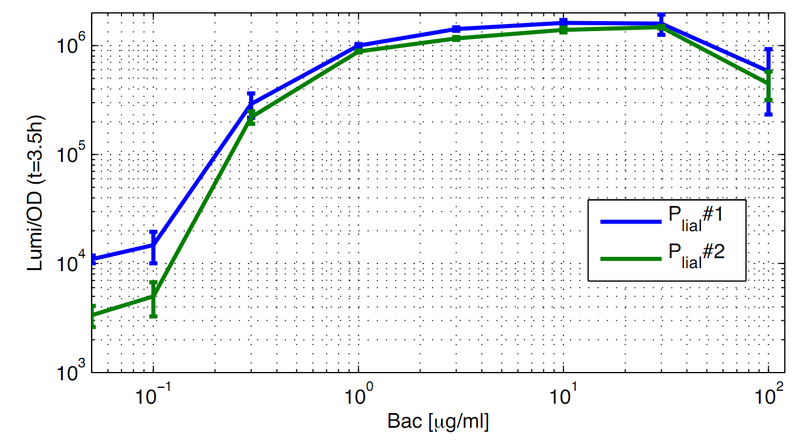
For better demonstration of the PliaI activity as a function of the bacitracin concentration, data from one timepoint at the experiment (Fig. 1) are shown depending on the bacitracin concentration (Fig. 2). Therefore Lumi/OD600 values are shown for the time point t=3,5h.
β-galactosidase assay
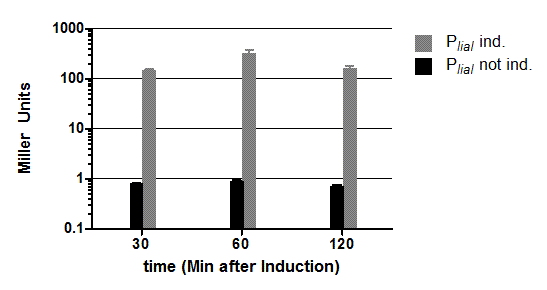
The inducible promoter PliaI was also evaluated with the reporter vector pSBBs1C-lacZ. (Fig.3).
|
Promoter activity leads to the expression of the β-galactosidase. The β-galactosidase assay of the inducible Bacillus promoter PliaI was repeated three times. We induced with bacitracin (20μg/ml) when the cultures reached an OD600 of 0.4. PliaI does not show any activity without induction. After induction with bacitracin there is a strong promoter activity that the produced enzyme reaches an activity of about 500 Miller Units. Summing up, the promoter activity after induction is about 500 times higher than without induction.
Sequence and Features
This part was amplified from the genome of B. subtilis.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]