Part:BBa_K823025
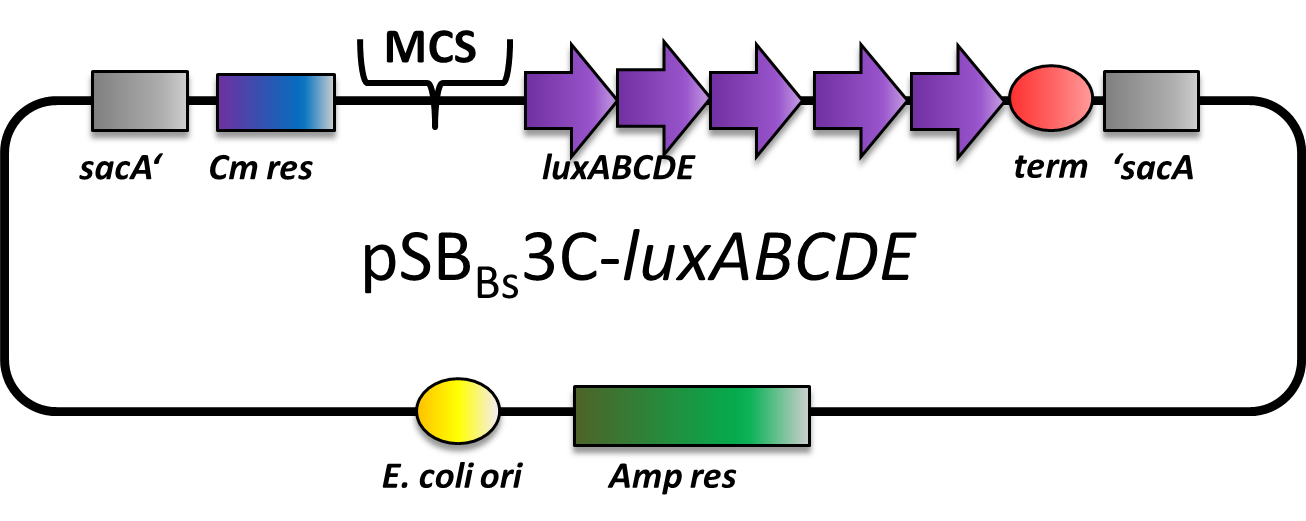
pSBBs3C-luxABCDE (lux reporter vector for B.subtilis)
This part is a reporter vector for Bacillus subtilis. It integrates at the sacA locus of B.subtilis genome. It has a ampicillin resistance for cloning in E.coli and Chloramphenicol resistance for selection in B. subtilis. The multiple cloning site contains an rfp-cassette for selection and is followed by the luxABCDE gene cluster for luminescence reporter assays.
This backbone is a BioBricked version of the B. subtilis vector pAH328. This part was also evaluated in the publication [http://www.jbioleng.org/content/7/1/29 The Bacillus BioBrick Box: generation and evaluation of essential genetic building blocks for standardized work with Bacillus subtilis] by Radeck et al..
This BioBrick is part of the LMU 2012 igem project [http://2012.igem.org/Team:LMU-Munich/Bacillus_BioBricks BacillusBioBrickBox]
Reference: [http://www.ncbi.nlm.nih.gov/pubmed/20709900 Schmalisch et al.]
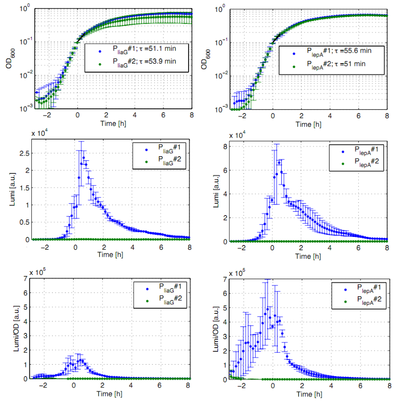
Fig. 1: Luminescence measurement of the constitutive Bacillus promoters PliaG and PlepA in the reporter vector pSBBs3C-luxABCDE. OD600 (up), LUMI (middle) and LUMI per OD600 (down) depending on the time (h) are shown for two different clones (green/blue). Data derive from three independent experiments, the graphs show the mean fo the three experiments and the standard deviation. Curves were fitted over each other (t=0, OD600=0,3) and smoothed by taking the average of three neighboring values.
For handling of B. subtilis vectors, please see here.
The transformation into B. subtilis is explained here.
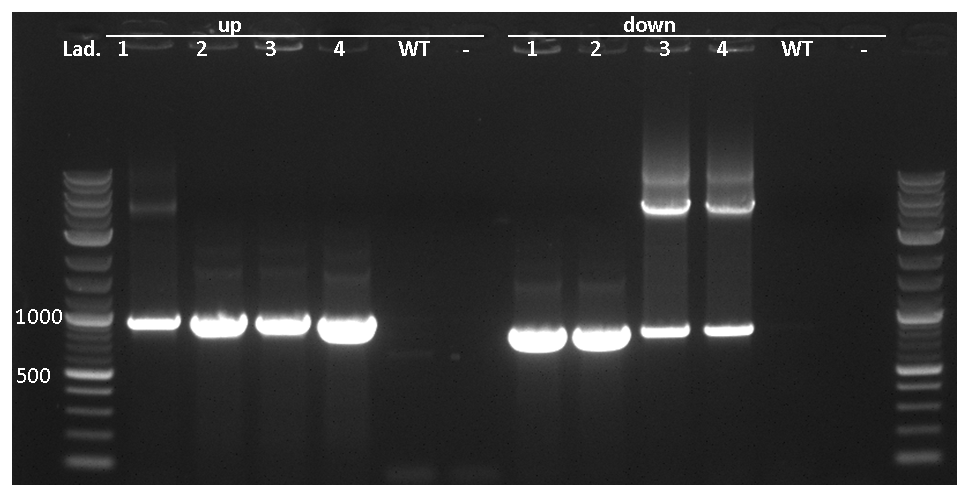
This vector integrates into the sacA locus of B. subtilis which is tested for by colony PCR (TM2505+06 and TM2507+08). One primer is located outside of the region and its respective partner is located on the inegrative part of the vector, facing outwards. gDNA from W168 as a negative control should not give a PCR product.
The constitutive promoters PliaG and PlepA were evaluated in the reporter vector pSBBs3C-luxABCDE which contains the lux operon. This is why the promoter activity leads to gene expression and to the production of the protein luciferase. The luminescence of this protein can be measured with the plate reader Synergy2 (Biotek) (Fig.1). All clones show a normal growth behaviour. The activity of the promoters increases during transition from log to stationary phase. The second clone of the promoters PlepA and PliaG did not show any luminescence activity. Therefore additional clones should be measured. In the beginning of the growth curve the activity of both promoters increases to their maximum. This maximum activity appears during the same growth phase. PliaG has an activity maximum of about 100.000 Lumi/OD600 during the transition from logarithmic to the stationary phase. PlepA shows a maximum of about 400.000 Lumi/OD600. Comparing these two constitutive promoters the activity of PlepA is about four times higher than the activity of PliaG. In the late stationary phase the activity completely disappears.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 10619
Illegal SpeI site found at 2
Illegal PstI site found at 16
Illegal NotI site found at 9
Illegal NotI site found at 10625 - 21INCOMPATIBLE WITH RFC[21]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal EcoRI site found at 10619
Illegal BglII site found at 5170
Illegal BamHI site found at 5705 - 23INCOMPATIBLE WITH RFC[23]Illegal prefix found at 10619
Illegal suffix found at 2 - 25INCOMPATIBLE WITH RFC[25]Illegal prefix found at 10619
Plasmid lacks a suffix.
Illegal XbaI site found at 10634
Illegal SpeI site found at 2
Illegal PstI site found at 16
Illegal NgoMIV site found at 9536
Illegal AgeI site found at 576 - 1000INCOMPATIBLE WITH RFC[1000]Plasmid lacks a prefix.
Plasmid lacks a suffix.
Illegal BsaI site found at 7576
Illegal BsaI.rc site found at 3489
Illegal SapI site found at 1095
Illegal SapI.rc site found at 8658
//chassis/prokaryote/bsubtilis
//chassis/prokaryote/ecoli
//plasmid/chromosomalintegration
//plasmidbackbone/libraryscreening/promoter
| chassis | B. subtilis + E. coli |
| copies | integrative in B. subtilis |
| origin | pAH328 |
| resistance | A(E. coli) + C(B. subtilis) |