Difference between revisions of "Part:BBa K822004"
| Line 6: | Line 6: | ||
This part is a duplicate of <partinfo>BBa_K292006</partinfo>. The NTNU iGEM 2011 team found that the physical DNA available from the registry for that part [https://parts.igem.org/Part:BBa_K292006:Experience did not match the registered sequence] , and we have therefore remade it. Sequencing confirms that this part has the desired sequence. | This part is a duplicate of <partinfo>BBa_K292006</partinfo>. The NTNU iGEM 2011 team found that the physical DNA available from the registry for that part [https://parts.igem.org/Part:BBa_K292006:Experience did not match the registered sequence] , and we have therefore remade it. Sequencing confirms that this part has the desired sequence. | ||
| + | {|border="0" | ||
| + | |[[image:Testkutt_BBa_K292006.png|300px]] | ||
| + | |[[image:RBS+LacI+term-gel.PNG|300px]] | ||
| + | |- | ||
| + | |This is the test cut of BBa_K292006 that the NTNU iGEM team 2011 performed. The testcut was performed with EcoRI+PstI (expected fragments: 1303 bp + 2053 bp), BglI+BclI (expected fragments: 1324 bp + 2032 bp), BglI+EcoRV (expected fragments: 1596 bp + 1760 bp) and BglI+BanII (expected fragments: 1521 bp + 1835 bp). The test cut shows that none of the expected fragments are present. | ||
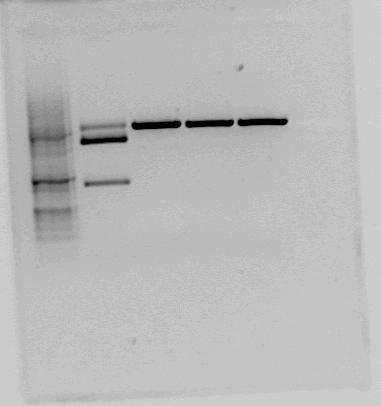
| + | |Test cut of our improved part performed with NotI (first red box, expected fragments: 1276 bp + 2055 bp) and XbaI+PstI (second red box, expected fragments: 1278 bp + 2053 bp). The fragments cut with NotI makes sense on gel. In the case of cutting with XbaI+PstI, we did not expect three fragments, but the upper fragment could be uncut plasmid, since the lower fragments makes sense. | ||
| + | |} | ||
| + | |||
| + | The sequencing results can be found [http://2012.igem.org/Team:NTNU_Trondheim/Sequencing_Improved_Construct here]. The sequencing result shows that in the old biobrick, only the terminator is present, and no LacI or RBS. In our improved biobrick, both RBS, LacI and terminator are present. | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 21:36, 26 September 2012
LacI generator (no promoter)
A composite part with coding sequence for LacI-LVA together with ribosome binding site (RBS) and a double transcriptional terminator. For use in regulation of LacI-repressed genetic elements, by combining the part with an appropriate promoter.
This part is a duplicate of BBa_K292006. The NTNU iGEM 2011 team found that the physical DNA available from the registry for that part did not match the registered sequence , and we have therefore remade it. Sequencing confirms that this part has the desired sequence.
The sequencing results can be found [http://2012.igem.org/Team:NTNU_Trondheim/Sequencing_Improved_Construct here]. The sequencing result shows that in the old biobrick, only the terminator is present, and no LacI or RBS. In our improved biobrick, both RBS, LacI and terminator are present.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1168
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]