Part:BBa_K801042
GAL4 based yeast light-switchable promoter system
composite part of Bba_K319003, BBa_K801039, BBa_K801011 Bba_K319003, Bba_K801040, and BBa_K801011
Background and principles
This system bases on the yeast two-hybrid system which was originally created for exploring protein-protein interactions. One candidate of a potential protein-interaction pair is fused to the DNA-binding domain of a transcription factor and the other candidate to the activation domain of a transcription factor. If the proteins candidates are really physically interacting with each other, this event will starts the transcription of downstream reporter genes, e. g. Luciferase, LacZ or an auxotrophic marker.
Reverse yeast-two hybrid based light-switchable promoter system
This basic principle is utilized in the yeast light-switchable promoter system. But in contrast to yeast-two hybrid, we already know the interaction partners (PhyB and PIF3). The photoconvertible binding of PhyB to PIF3 is used, to recover the physical contiguity of the DNA binding domain and the transcriptional activation domain under defined conditions (red light).
This light-inducible system contains two proteins, phytochrome B (PhyB) and phytochrome interacting factor 3 (PIF3). PhyB and PIF3 will just form a heterodimer, if PhyB is exposed to red light. Exposition under red light leads to a conformation change of PhyB to its active form (Pfr-form); the Pfr form of PhyB now can bind PIF3. PhyB comprises a light-absorbing chromophore phycocyanobilin, which gives PhyB the ability to undergo a photoconversion to the active Pfr form (red light exposition) or back to its ground-state Pr (far-red light exposition or darkness).
GAL4 based light-switchable promoter system
In our first case we create two constitutively expressed fusion proteins, the first one is PhyB fused to GAL4DBD for the DNA binding part (BBa_K801040 and the second one is PIF3 fused to GAL4AD for the transcriptional activating part (BBa_K801039). This system allows us to control spatio-temporally the expression of our genes coded on pTUM104 and driven by the GAL1 promoter (The TATA-box of pGAL1 is preceded by binding elements for GAL4). To prevent interference with the endogenous GAL4 system of yeast, we are using the Y190 S. cerevisiae strain, which has a GAL4/GAL80 deletion.
One great advantage of the GAL4 based system is that we can use all our constructs which we have first cloned downstream of a GAL1 promoter without further cloning steps! But the disadvantage is that we have to use a yeast strain carrying a GAL4/GAL80 deletion.
If you want to use a supermarket yeast or a brewing strain you have to use the LexA based light-switchable promoter system, described in the next section.
LexA based light-switchable-promoter system
For more information about the LexA based system, please see here: BBa_K801043
Characterisation via Luciferase Assay
Induction Setup
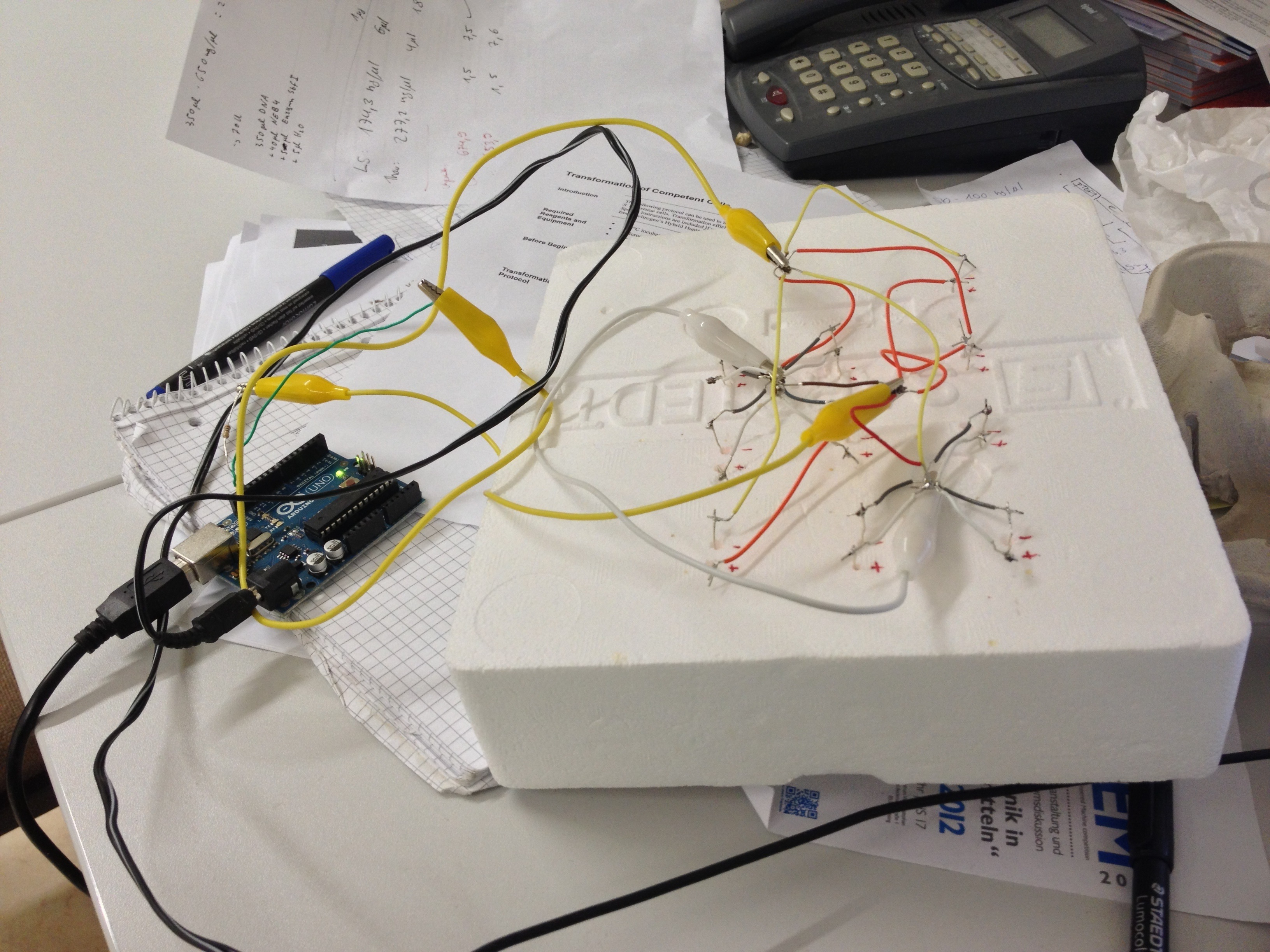
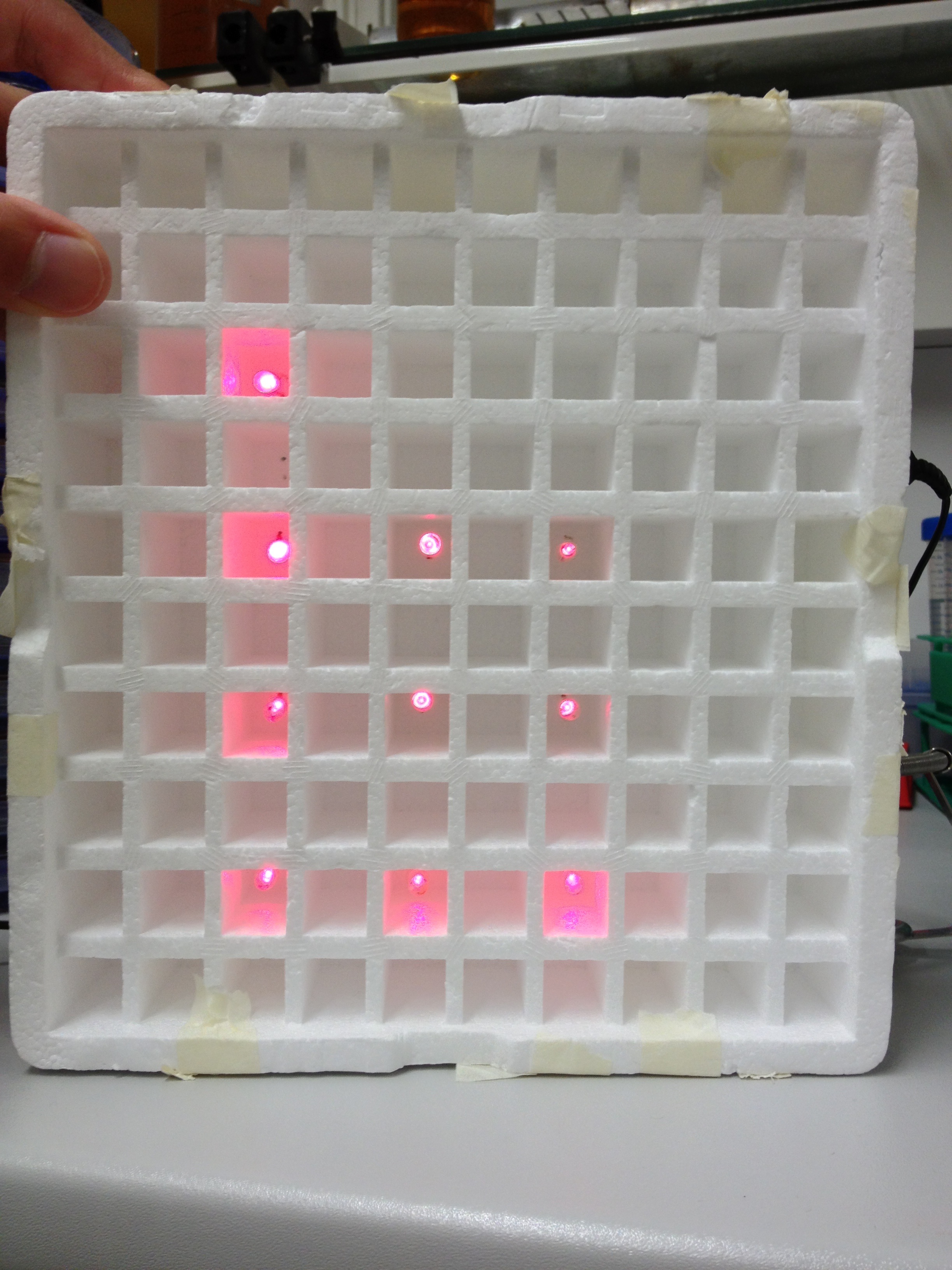
An array of 10 LEDs with emission peak at 660 nm ([http://www.alldatasheet.com/datasheet-pdf/pdf/296270/ROITHNER/B5-436-30D.html Data sheet]) were attached into the molds of the packaging of 2 ml cuvettes and soldered together on the rear side of the packaging. As the cuvettes are the very ones that will later be used for illumination of the cells, the use of the packaging as LED matrix will allow quick removal during measurements and enhance accuracy of results.
Literature suggest pulsed illumination of the cells with a pulse duration of 10 seconds and a pulse frequency of 1 pulse every 5 minutes. The LEDs are actuated with an Arduino UNO micro-controller that puts the suggested protocol. The use of a micro-controller will allow us to easily test different pulse lengths and frequencies.
Results
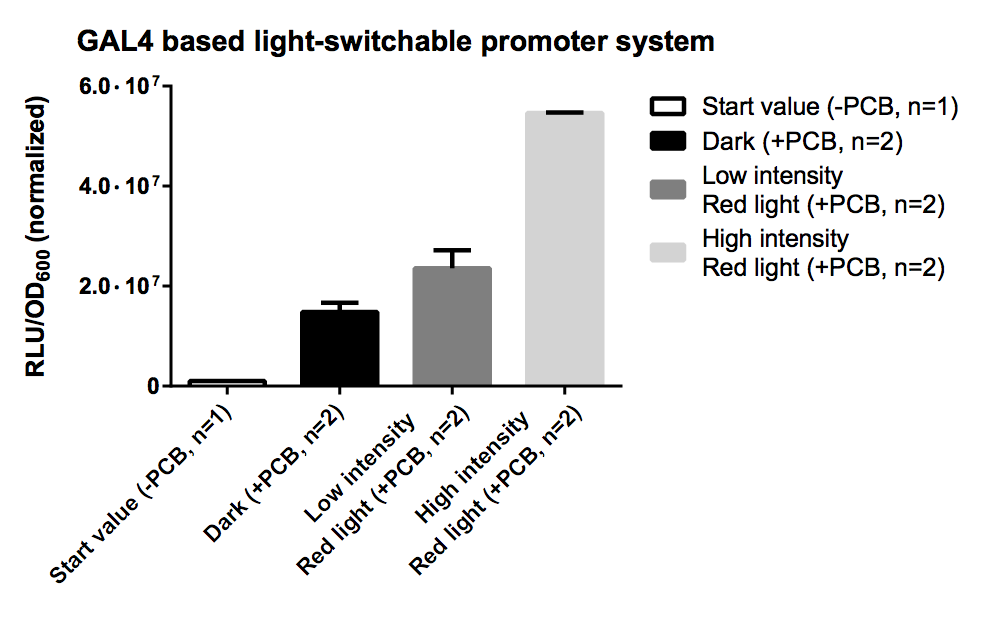
PCB is necessary for correct folding of the PhyB domain, hence without PCB the output of our reporter system is close to zero.
The sample irradiated with high intensity of red light shows a nearly 4-fold increasement in gene expression for Renilla luciferase compared to the sample illuminated with low intensity (10% of high intensity) of red light.
Expression of both fusion proteins were driven by the strong pTEF1 promoter and the gene expression battery was cloned on a high copy shuttle vector, which leads to a quite high level of both fusion proteins. This will result in a significant basal activity in gene expression of reporter genes due to unspecific binding of both fusion proteins. This phenomenon could be easily avoided by using a low copy shuttle vectors or by integrate this casette into the genome to get a high S/N ratio.
Recommendation for teams using this part
Using a low copy shuttle vector or genome integration is recommended while using this part becuase both fusion proteins are already driven by a strong pTEF1 promoter.
The essential chromophore phycocyanobilin (PCB) can be isolated by [http://2012.igem.org/Team:TU_Munich/Notebook/Protocols methanolysis and chloroform extraction]. Biosynthesis of phycocyanoblin can be reached by simultaneously expressing heme oxygenase and phycocyanobilin:ferredoxin oxidoreductase, so no PCB addition is nescessary anymore.
If you want to add PCB to the medium, a total concentration between 5 µM-25 µM is recommended. PCB concentration can be calculated with the absorption value at 680 nm: c(in mM) = A680/37.9 x dilution factor.
References
- http://www.ncbi.nlm.nih.gov/pubmed/15823535 Chen et al., 2005 Chen, M., Tao, Y., Lim, J., Shaw, A., and Chory, J. (2005). Regulation of phytochrome B nuclear localization through light-dependent unmasking of nuclear-localization signals. Curr Biol, 15(7):637–42.
- http://www.ncbi.nlm.nih.gov/pubmed/19165330 Kikis et al., 2009 Kikis, E. A., Oka, Y., Hudson, M. E., Nagatani, A., and Quail, P. H. (2009). Residues clustered in the light-sensing knot of phytochrome B are necessary for conformer-specific binding to signaling partner PIF3. PLoS Genet, 5(1):e1000352.
- http://www.ncbi.nlm.nih.gov/pubmed/19749742 Levskaya et al., 2009 Levskaya, A., Weiner, O. D., Lim, W. A., and Voigt, C. A. (2009). Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature, 461(7266):997–1001.
- http://www.ncbi.nlm.nih.gov/pubmed/12355112 Mendelsohn, 2002 Mendelsohn, A. R. (2002). An enlightened genetic switch. Nat Biotechnol, 20(10):985–7.
- http://www.ncbi.nlm.nih.gov/pubmed/12219076 Shimizu-Sato et al., 2002 Shimizu-Sato, S., Huq, E., Tepperman, J. M., and Quail, P. H. (2002). A light-switchable gene promoter system. Nat Biotechnol, 20(10):1041–4.
- http://www.ncbi.nlm.nih.gov/pubmed/15486100 Khanna et al., 2004 Khanna, R., Huq, E., Kikis, E. A., Al-Sady, B., Lanzatella, C., and Quail, P. H. (2004). A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell, 16(11):3033–44.
- http://www.ncbi.nlm.nih.gov/pubmed/11553807 Gambetta and Lagarias, 2001 Gambetta, G. A. and Lagarias, J. C. (2001). Genetic engineering of phytochrome biosynthesis in bacteria. Proc Natl Acad Sci U S A, 98(19):10566–71.
- http://www.ncbi.nlm.nih.gov/pubmed/10466729 Ni et al., 1999 Ni, M., Tepperman, J. M., and Quail, P. H. (1999). Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature, 400(6746):781–4.
- http://www.ncbi.nlm.nih.gov/pubmed/12734586 Van Criekinge and Beyaert, 1999 Van Criekinge, W. and Beyaert, R. (1999). Yeast two-hybrid: State of the art. Biol Proced Online, 2:1–38.
- http://www.ncbi.nlm.nih.gov/pubmed/3891738 Wertman and Mount, 1985 Wertman, K. F. and Mount, D. W. (1985). Nucleotide sequence binding specificity of the LexA repressor of Escherichia coli K-12. J Bacteriol, 163(1):376–84.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 862
Illegal BglII site found at 2795
Illegal BamHI site found at 2877
Illegal XhoI site found at 2828
Illegal XhoI site found at 2847
Illegal XhoI site found at 4982
Illegal XhoI site found at 5316 - 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 2240
Illegal AgeI site found at 5540 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 4367
Illegal BsaI site found at 4773
Illegal BsaI site found at 5235
Illegal BsaI.rc site found at 205
Illegal BsaI.rc site found at 1986
Illegal SapI site found at 3044
//promoter
//proteindomain
//proteindomain/activation
//proteindomain/binding
//proteindomain/dnabinding
//proteindomain/localization
| None |