Difference between revisions of "Part:BBa K729001"
Sednanalien (Talk | contribs) (→Results) |
(→Results) |
||
| Line 34: | Line 34: | ||
[[Image:UniversityCollegeLondon_IrrE_Growth_Curves_LB.png|470px]] | [[Image:UniversityCollegeLondon_IrrE_Growth_Curves_LB.png|470px]] | ||
[[Image:UniversityCollegeLondon_IrrE_Growth_Curves_LB%2B0.3.png|470px]] | [[Image:UniversityCollegeLondon_IrrE_Growth_Curves_LB%2B0.3.png|470px]] | ||
| − | [[Image:UniversityCollegeLondon_IrrE_Growth_Curves_LB%2B0.6.png|470px]] | + | [[Image:UniversityCollegeLondon_IrrE_Growth_Curves_LB%2B0.6.png|470px|centre]] |
===Modelling=== | ===Modelling=== | ||
Revision as of 15:31, 21 September 2012
irrE from Deinococcus radiodurans
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Description
The protein IrrE originates from Deinococcus radiodurans, where it confers resistance to radiation. When transformed into E. Coli however, it protected against salt, oxidative and thermal shock. IrrE appears to function as a global regulator of stress factor genes. So far it has been demonstrated to upregulate transcription of recA and pprA – genes which encode Recombinase A and Radiation Inducible Protein. With respect to salt tolerance, IrrE upregulates the production of several stress responsive proteins, protein kinases, metabolic proteins, and detoxification proteins. It also downregulates glycerol degradation. With this global regulatory effect, E. Coli becomes more salt tolerant.
In order to characterise this BioBrick, we ligated it with the Constitutive Promoter BBa_J23119, and Ribosome Binding Site BBa_B0034. This produced the final construct, BBa_K729005.
Characterisation
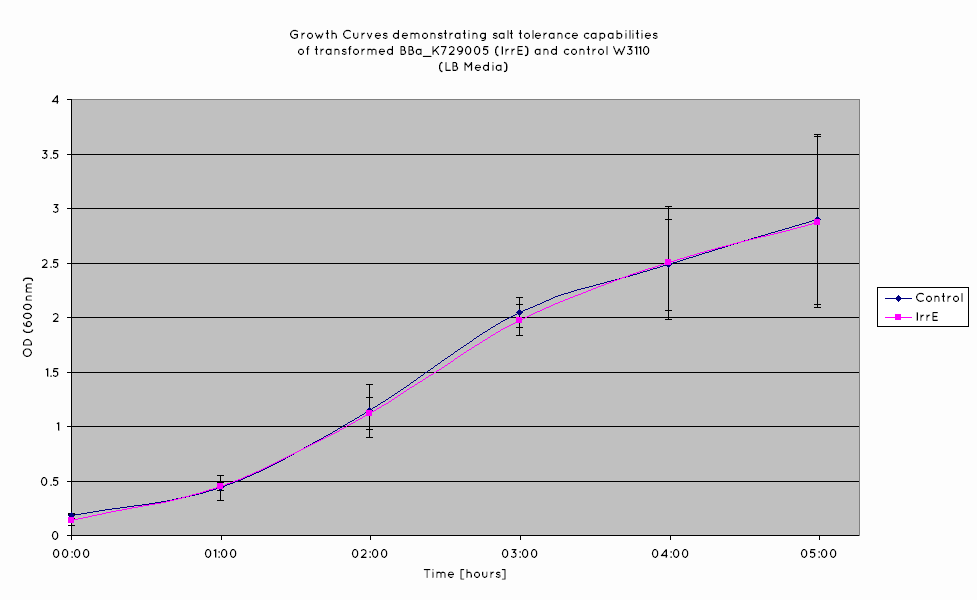
IrrE is deduced to globally regulate gene expression in E. Coli, and to date, no specific assay allows for the convenient determination of IrrE expression. As such, the expression of IrrE will be determined indirectly by determining the increased salt tolerance of our cells. In order to confirm this, we will be conducting growth curve experiments. This will allow us to determine if our cell grow to a higher density in high salinity when compared to the untransformed cells.
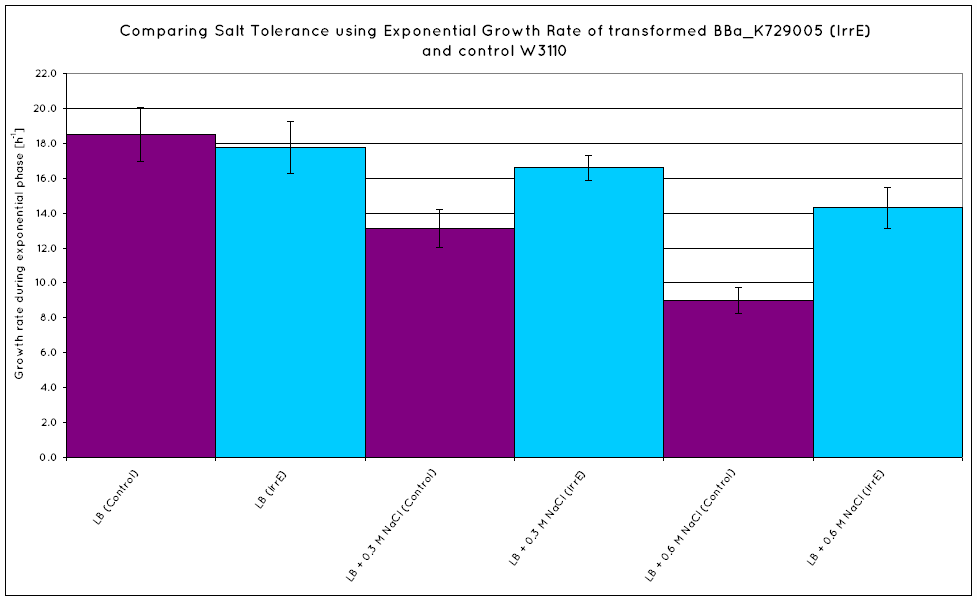
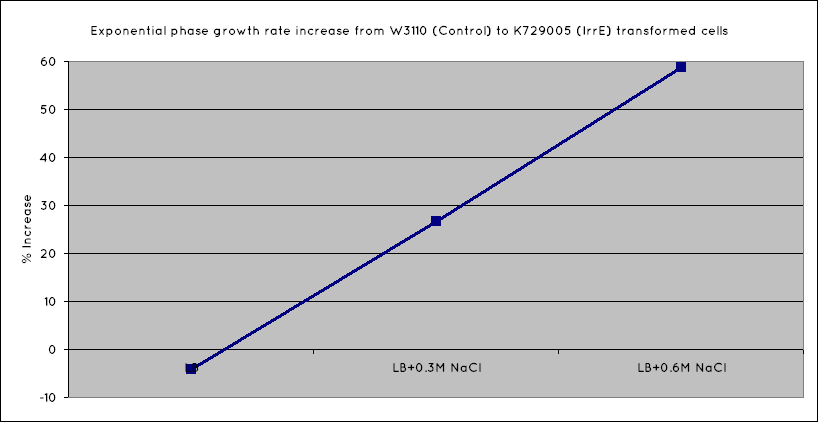
Results
From our results, we observed that E. Coli transformed with the IrrE gene exhibit greater salt tolerance than their non-transformed counterparts. This is revealed in the greater OD the cells grow to in high salt media, as well as their increased growth rate. Our results are consistent with the results obtained in the original paper (Pan et al. 2009), suggesting that our cells are indeed expressing the IrrE global regulator.
Modelling
Conclusion
We have determined that IrrE is an ideal global regulatory gene to be utilised in conferring salt tolerance on our cells. Transforming our cells with the IrrE gene, we have successfully increase the growth rate of E. Coli in high salinity conditions.
References
"IrrE, a global regulator of extreme radiation resistance in Deinococcus radiodurans, enhances salt tolerance in Escherichia coli and Brassica napus.(Pan, J., Wang, J., Zhou, Z.(2009)PloS one, 4(2), e4422.)