Part:BBa_K4491007
pBAD_AP
| pBAD_AP | |
|---|---|
| Function | Inducible promoter |
| Chassis Tested | Escherichia coli (bacterial) |
| Assembly Used | JUMP Assembly |
| Abstraction Hierarchy | Part |
| Related Device | BBa_K2442101 |
| Backbone | pSC101 |
| Submitted by | Cambridge iGEM 2022 |
This gene encodes an improved version of the wild-type araBAD promoter called pBAD_AP2, an inducible promoter controlled by araC protein. The native pBAD is part of the araBAD operon, which is responsible for regulating arabinose metabolism in E.coli. pBAD_AP2 belongs to a collection of newly-designed araBAD promoters that are below 160 bp! As the Registry does not generally allow an entry with multiple sequences, we gave the sequence of pBAD_AP2 only as a representative (shows significant improvement), but all remaining sequences are also given below for completeness .
Contents
Usage and Biology
The exact definition of the araBAD promoter varies ambiguously between sources. Historically, pBAD only refers to a short segment upstream of the +1 transcription start site (referred to as the core promoter), containing the -35 and -10 boxes. However, as a complete promoter Part, the regulatory region further upstream is also included. In the following discussion, our definition of pBAD refers to the whole sequence consisting of both the regulatory sequence and the core promoter.
The araBAD promoter regulates the araBAD operon and is controlled by araC - a regulatory protein known for its “love-hate” mechanism of action. The upstream regulatory region consists of various protein binding sites - araI1, araI2, araO1 and araO2 can be occupied by araC. Between araI1 and araO1 also lies a CAP binding site, which recruits Catabolite receptor protein for transcription activation. The spacing between araO1 and O2 is noticeably large, reaching nearly 210 bp, which, unsurprisingly, is responsible for the overall bulkiness of the promoter. Interestingly, a region within this spacer contains a promoter of araC gene, which runs in the opposite direction to the araBAD operon. This promoter is regulated by araO1 just downstream. The araC protein therefore not only regulates the araBAD operon but also controls its own production by binding to araO1 region (negative autoregulation).
In the absence of L-arabinose, transcription is repressed by the action of two araC molecules binding to the structure. One araC binds to araI1 and the other binds to araO2 further upstream. The dimerization domain confers high affinity, bringing the two protein molecules closer to dimerize, essentially creating a DNA loop as a result. This looping mechanism prevents any sigma factors, RNA Polymerase or CRP from being recruited, thus repressing transcription.
In the presence of L-arabinose, binding of the sugar molecule to the arabinose-binding domain triggers a conformational change in the DNA-binding domain of araC, which reduces its affinity to distal araO2 site. This makes the protein more favourable to bind to araI2, just downstream of araI1. Therefore, the dimerized complex is formed at araI1-araI2 site instead of araO2-araI1, which breaks the loop and hence activates transcription. The dimer araC also “nudges” the RNA Polymerase for enhanced transcription.
pBAD Entries in the Registry
Our team essentially created a family of combinatorial pBADs, some of which yielded significant improvement from the wild-type version. We list below several previous entries of the araBAD promoters, some of which may offer stronger merits in certain aspects. All of our designs have a truncated length of less than 160 base pairs, which is only half of the typical size.
- BBa_K2442101 by team Glasgow 2017
- PBAD_SPL by team Denmark DTU 2013.
- PBAD_Promoter_Family by team British Columbia 2009.
Preamble from the Cambridge team
The Cambridge 2022 Team had intended to use the araBAD promoter to build an antithetic feedback circuit that demonstrates robust perfect adaptation (RPA). However, we were concerned that wild-type PBAD is relatively large, topping at around 300 bp, which increases the bulkiness of our Level-2 construct and reduces efficiency. Also, pBAD, by nature, is a relatively leaky and medium-strength promoter, therefore, lots of araC is needed to fully reach maximal activation. However, overexpression of araC is toxic to cells, and thus can drastically impede performance of the circuit. we therefore thought it would be an interesting Part Improvement project to redesign the existing wild-type promoter. Our initial goal was to engineer a PBAD with minimal length, but shows both lower leakiness and higher maximal activity. To achieve this, we did intensive literature search for prior optimization strategies. To understand the rationales, we also gathered information on the regulatory mechanism of the promoter, which was presumably only prevalent in papers from the 1970s.
We envisioned that the significant reduction in the promoter’s size can be of frugal advantage for future circuit designs. With less than 150 bp, the promoter can be synthesised by oligo-annealing, which is significantly faster and cheaper than ordering DNA parts. This could also be of great benefit for future iGEM teams from regions where DNA synthesis and delivery are often slow, allowing them to speed up project timelines.
pBAD_AP sequences
The table below shows nine newly-designed araBAD promoters, all with less than 160 base pairs. To see our design rationales for all sequences, please check out the Design page.
| Identifiera | Rationale 1b | Rationale 2 | Rationale 3 | Part sequences | |
|---|---|---|---|---|---|
| AP1 | + | - | - | agaaaccaattgtccataattgattatttgcacggcgtcacactttgctatgccatagcatttttatccataagattagcggatccta
cctgacgctttttatcgcaactctctactgtttctccatacccg | |
| AP2 | + | + | - | agaaaccaattgtccataattgattatttgcacggcgtcacactttgctatgccatagcaagatagtccataagattagcgtttttat
cctgacgctttttatcgcaactctctactgtttctccatacccg | |
| AP3 | + | - | + | agaaaccaattgtccataattgattatttgcacggcgtcacactttgctatgccatagcatttttatccataagattagcggat
cctacctgacgctttttatcgcaactctctactgtttctccatacccg | |
| AP4 | + | + | + | agaaaccaattgtccataattgattatttgcacggcgtcacactttgctatgccatagcaagatagtccataagattagcgtttttat
cctgacgtgcgcctgccgtccaaagtaatatccttacatacccg | |
| AP5 | + | + (78)c | + | agaaaccaattgtccatattgcatcagacattgccgtcacattgattatttgcacggcgtcacactttgctatgccatagcaagatagt
ccataagattagcgtttttatcctgacgtgcgcctgccgtccaaagtaatatccttacatacccg | |
| AP6 | - | + | - | acattgattatttgcacggcgtcacactttgctatgccatagcaagatagtccataagattagcgtttttat
cctgacgctttttatcgcaactctctactgtttctccatacccg | |
| AP7 | - | + | + | acattgattatttgcacggcgtcacactttgctatgccatagcaagatagtccataagattagcgtttttat
cctgacgtgcgcctgccgtccaaagtaatatccttacatacccg | |
| AP8 | - | - | + | acattgattatttgcacggcgtcacactttgctatgccatagcatttttatccataagattagcggatccta
cctgacgtgcgcctgccgtccaaagtaatatccttacatacccg | |
| AP9 | - | - | - | agaaaccaattgtccataattgattatttgcacggcgtcacactttgctatgccatagcatttttatccataagattagcggatccta
cctgacgctttttatcgcaactctctactgtttctccatacccg | |
a AP refers to the initials of the principal designer of the araBAD promoters.
b(-) in Rationale 1 refers to complete removal of both araO2 and the spacing, but still leaves the CAP binding site.
c AP5 has a spacing of 78 bp instead of the common 56 bp.
Experimental design
To test the designed araBAD promoters, we cloned nine respective Level 0 promoter parts into Level 1 JUMP constructs, using a standard Golden Gate Assembly protocol with BsaI-HFv2. Lying downstream of the promoter is a B0032 medium RBS, an mVenus reporter and a DT5 double terminator. A medium-strength RBS was chosen to avoid burden caused by accidental overexpression of mVenus. All TUs were put into a pJUMP27-1A destination vector. The low copy number plasmid makes it favourable for avoiding phototoxicity and aggregate bodies, as well as reducing noise. A list of constructs ID and their respective constituent promoter is given below.
| LVL1 ID | Promoter |
|---|---|
| PB1 | AP1 |
| PB2 | AP2 |
| PB3 | AP3 |
| PB4 | AP4 |
| PB5 | AP5 |
| PB6 | AP6 |
| PB7 | AP7 |
| PB8 | AP8 |
| PB9 | AP9 |
| PB10 | BBa_K2442101 |
| PBneg | dummy control |
Golden-gate-assembled mixtures were chemically transformed into DH5alpha cells for selection on Kanamycin plates. cPCR was carried out on seemingly-good colonies to screen for bands with correct size. Colonies with successful assemblies, indicated by cPCR, were liquid cultured for next-day miniprep. The retrieved plasmids, once verified through Sanger sequencing, were then transformed into a Marionette-Wild strain by electroporation. We chose Marionette as the testing strain because it is highly optimised as an arabinose sensor. Colonies of Marionette cells (selected on Kanamycin plates) were inoculated in 2 mL Neidhardt EZ Rich Defined Medium (EZRDM) and shaking-incubated overnight for 20 hours, at 37oC and 250 rpm.
We conducted two experiments to validate the nine pBADs against the wild-type counterpart and a negative control. The negative control (termed PBneg) was a dummy construct with similar length, containing mVenus CDS, but no araBAD promoters. Both the wild-type (PB10) and the PBneg constructs were cloned using the method described above.
Both experiments would involve measuring mVenus fluorescence intensity (FI) of transformed Marionettes grown under varying arabinose concentrations. FI was measured using a ClarioStar plate reader, on Greiner F-bottom 96-well plates or standard Greiner 384-well plates.
Results
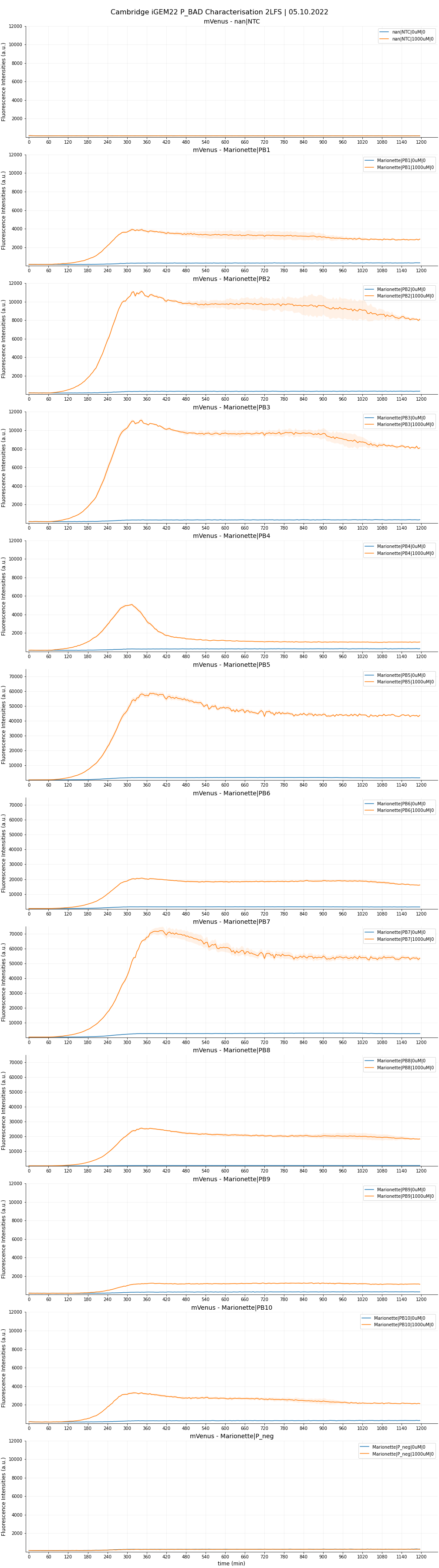
Two-Level factorial screen
We first investigated the leakiness and maximal strength of the eight P_BADs. We chose arabinose concentrations of 0 uM and 1000 uM as the testing conditions. We shall, for now, not take Hill’s dynamics into account, and focus solely on the two concentration endpoints that best showcase the promoters’ two characteristics. We also calculated the fold-change (or fold-induction), defined as the ratio of maximal expression to leakiness:
Fold change = Max expression / Leakiness
Each sample was done in four replicates. The raw data are plotted in Figure 5,6 and 7.
Hill's induction assay
We also hoped to characterise classical parameters of inducible systems, namely, the dissociation constant KD and the Hill’s coefficient n. Therefore, we carried out a combinatorial induction assay with arabinose concentrations ranging from 0, 10, 25, 50, 75, 100, 125, 200, 500, 1000 and 2000 uM. For this testing, we used a 384-well plate instead to test all constructs; and a robot called Opentron OT-2 to automate plate preparation. Results are shown below.
We selected PB1, PB2, PB3, PB8, PB9 and PB10 and used the template equation given in Meyer et al (2019) for least-square regression. An example plot for the fitted curve of PB10 is given in Figure 10, and the six fitted functions are collected in a single plot in Figure 11.
| LVL1 ID | KD (uM) (fitted) | Hill's coefficient n (fitted) |
|---|---|---|
| PB1 | 29.6 | 0.54 |
| PB2 | 31.1 | 0.56 |
| PB3 | 32.9 | 0.54 |
| PB8 | 64.5 | 0.63 |
| PB9 | 140 | 0.67 |
| PB10 | 179 | 1.25 |
Discussion and outlook
Result from the 2LF-screen shows that PB1, PB2, PB3 and PB8 constructs (corresponding to promoter AP1, AP2, AP3 and AP8) showcased higher maximal expression but maintained same level of leakiness as that of PB10 (wild-type pBAD). AP2 and AP3 are definite contenders for significant improvements, being only half the length of their wild-type counterpart. We do not consider here AP8 as an improvement in our project, as this single rationale was fully accredited by the 2013 DTU Team. Still, it is worthwhile to note that in their previous characterization, the SPL (-35)-to-(-10) segment was incorporated into the wildtype length. Here, we put the optimised segment into the truncated 130 bp length, which also showed predictably high fold-change. We had slight issues with the induction assay. The experimental design failed to take into account the arabinose concentration range between 0 and 10uM. Therefore, for PB5, PB6, and PB7, we could not investigate the kinetics within this range. Our preliminary fitted curves for these functions (not shown) indicated a very steep, ‘all-or-nothing’ behaviour, and their maximal expression starts to plateau from even as low as 10uM. We hypothesised such observations with two possibilities:
- These promoters were ‘accidentally’ engineered to have a very sharp dynamic range (ie. Hill’s coefficient is very large) and thus confer a “switch-like” mechanism of action. In other words, only a minimal concentration of arabinose is enough to fully activate the promoter. (Note that this was not our original aim of improvement).
- These promoters still have predicted Hill’s kinetics, but the sigmoidal range (between 0 and 10uM) was not investigated thoroughly. This would mean that their dissociation constants are much smaller than that of the wild-type, shifting the functions left.
More careful testing must be done to verify these scenarios.
It was, perhaps not very surprising, that AP4 (containing all three optimizations) did not live up to the expectation. We hypothesised that the modification in araI sites interfered with the SPL segment, which severely reduces the maximal strength. We also observed the same phenomenon between AP6 (- + +) and AP8 (- - +). From the time-dependent FI curve, we observed that cells with PB4 construct (containing AP4) consistently showed a strong fluorescence burst, followed by a quick drop. Furthermore, beyond 10uM, PB4 shared similar behaviour with the three promoters above in that FI is relatively unchanged (possibly reaching maximum already). Glancing at OD600 graphs, we noted that PB4 cell viability is comparable to the rest, with no sign of impeded growth. We hypothesised that the FI drop could be due to an intrinsic mechanism that degrades mVenus protein, or somehow purposely interferes with the transcription mechanism. It might also be possible that the overengineered mechanism causes a ‘’congestion’’ of araC onto the araI1-araO2 site, which permanently blocks transcription. Though PB4 is unable to serve as an improved part, future investigations could generate very interesting knowledge about the underlying emergent mechanism of this P_BAD.
Moreover, AP5, which is similar to AP4 but instead with a 78 bp spacing, showed some promise for its towering maximal strength, but is unfortunately devalued by the higher leakiness compared to wild-type. It is interesting to note that AP5 did not suffer the burst-effect like AP4 did; therefore, it can be reasonably assumed that the spacing of 56 bp did have a drastic influence on AP4’s performance.
We are optimistic that AP9, despite lacking all three optimizations and thus being the most truncated, can be of use in certain circumstances where an inducible promoter with low expression is favoured. We therefore also made the sequence available on the Registry.
| dissociation_constant | 31.1 |
| hill_coeficient | 0.56 |