Part:BBa_K4391002
Salmonella typhimurium Aptamer
This is an high affinity ssDNA aptamer against bacteria Salmonella typhumurium . The aptamer 3D folded structure was generated, molecular docking against most of the known surface proteins of S. typhimurium was performed to identify the protein(s) the aptamer may bind to. Then the molecular dynamics simulation was performed.
Usage and Biology
Among a multitude of contagious enteric bacterial infections, salmonellosis is one of the most frequently reported bacterial foodborne diseases and is a major economic and public health concern worldwide. Of particular concern is salmonellosis caused by multidrug-resistant strains such as Salmonella enterica serovar Typhimurium. Therefore, the inspection of food for the presence of Salmonella has become routine all over the world.
Aptamers are single-stranded DNA or RNA oligonucleotides that bind target biomolecules with high affinity and specificity. There are several advantages to the use of aptamers as compared to antibodies: aptamers bind target molecules with affinity and specificity equal or superior to those of antibodies, they are easily synthesized and modified, and they are more stable than antibodies. Due to this studies on aptamers used as therapeutic, diagnostic, and analytical reagents have become more important. a Duan et. al. have selected the apatmer (ST2P) binding to Salmonella typhimurium by a whole- bacterium-based Systemic Evolution of Ligands by Exponential Enrichment (SELEX) procedure.
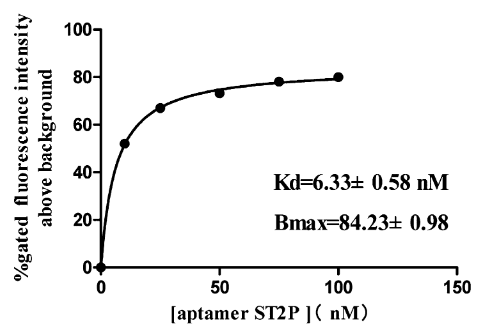
Fig-1 shows a typical binding saturation curve from flow cytometric analysis of the fluorescently labeled ST2P aptamer and the S. typhimurium cells used for selection (108 cfu/mL). Estimates from a nonlinear regression curve fit yield a Kd value of 6.33 ± 0.58 nM and a Bmax of 84% gated fluorescence above background. Binding of a fluorescently labeled randomized oligonucleotide library to the S. typhimurium cell mixture was also examined as a negative control.
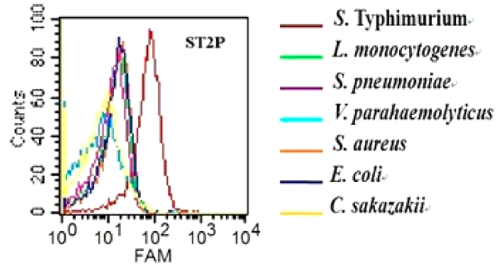
Characterization of specificity of ST2P aptamer for S. typhimurium was done by Duan et. al. using flow cytometry assay for the binding of the aptamer to 7 different bacteria. The 5′-FAM-labeled individual aptamers were incubated with 108 bacteria at 37 °C for 45 min. Fig-2 shows that selected aptamer sequence preferentially binds to S. typhimurium over the other cells tested.
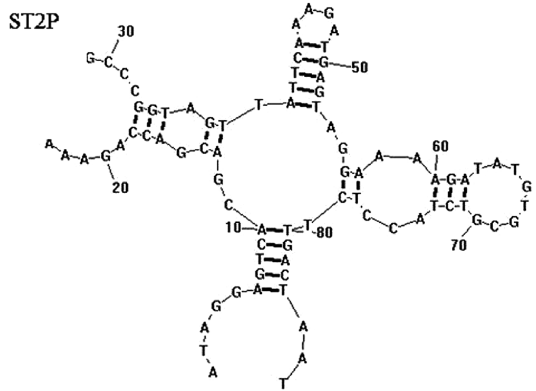
The secondary structure of the aptamer was predicted by Duan et. al. using RNA structure 3.0 with input conditions of room temperature (21 °C) and 1 mM MgCl. The most likely structure (Fig-3) was chosen on the basis of the lowest predicted free energy of formation (ΔG; kcal/mol).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Modelling
3D structure of the Aptamer
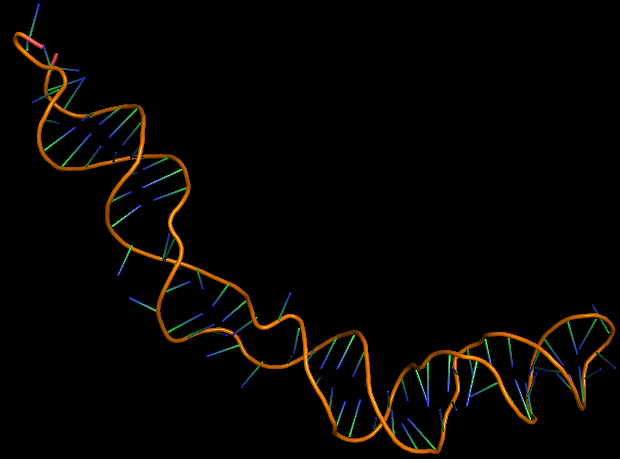
The 3D folded structure of the aptamer was generated using Xiao Lab 3dRNA/DNA - An RNA and DNA tertiary structure prediction method (http://biophy.hust.edu.cn/new/3dRNA/create). The input was the DNA aptamer sequence, molecule type DNA, Predict 2nd Structure: Yes, 2nd Structure Prediction Method: RNA Fold, Procedure Best, # of Predictions 5. No changes were made in Advanced options. Model-Fig-1 shows the most stable folded 3D structure.
Molecular Docking
Since the aptamer was selected by whole cell-SELEX, the exact macromolecule it binds to is not known. An attempt was in finding what exactly the apatmer binds to on S. typhimurium using molecular docking simulations. This model assumes that the aptamer binds to a membrane protein of S. typhimurium.
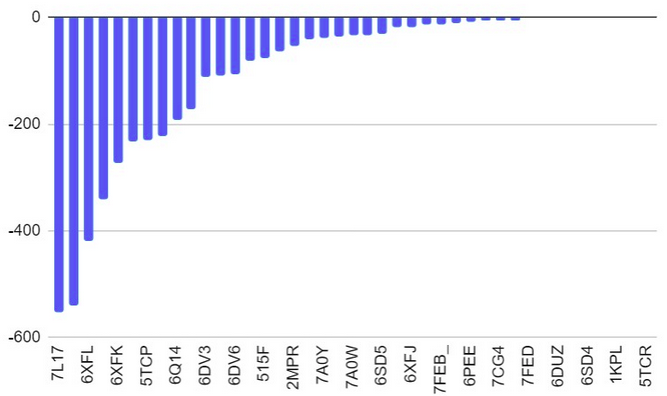
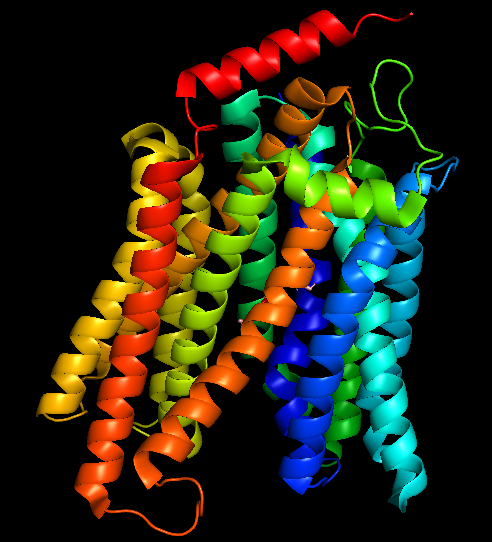
The surface/membrane proteins of S. typhimurium was identified from RSCB PDB. 68 total proteins were identified. Then each of these proteins were docked with the aptamer on Hex 8.0.0 software. Model-Fig-2 shows the binding energies of the most stable docking configurations of the proteins, obtained after docking. Protein melibiose permease with PDB ID 7L17 (Model-Fig-3) best docks with the aptamer (it has the least energy).
References
[1] Duan, Nuo & Wu, Shijia & Chen, Xiujuan & Huang, Yukun & Xia, Yu & Ma, Xiaoyuan & Wang, Zhouping. (2013). Selection and Characterization of Aptamers against Salmonella typhimurium Using Whole-Bacterium Systemic Evolution of Ligands by Exponential Enrichment (SELEX). Journal of agricultural and food chemistry. 61. 10.1021/jf400767d.
[2] http://hex.loria.fr/
[3] Zhao, Y., et al., Automated and fast building of three-dimensional RNA structures. Scientific Reports, 2012. 2: p. 734.
[4] Rcsb.org. H.M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T.N. Bhat, H. Weissig, I.N. Shindyalov, P.E. Bourne. (2000) The Protein Data Bank Nucleic Acids Research, 28: 235-242.
[5]
| None |